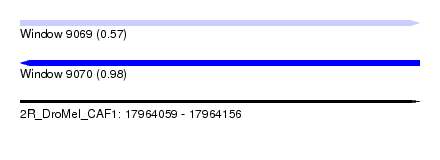
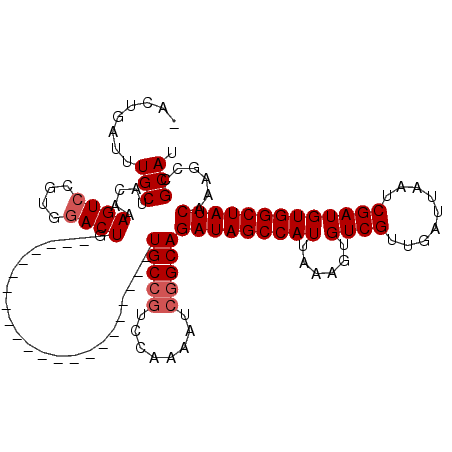
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,964,059 – 17,964,156 |
| Length | 97 |
| Max. P | 0.984660 |

| Location | 17,964,059 – 17,964,156 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.70 |
| Mean single sequence MFE | -27.91 |
| Consensus MFE | -21.87 |
| Energy contribution | -21.75 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.565719 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17964059 97 + 20766785 AUGCGGCUUUGAUAGCCACAUCGAUUAAUCAACGACACUUUAAUGGCUAUCUGCAGAUUUGGGUUGGCA----------------------CAGUCCACGGACUUUGUAGCAAAUCAGU- .....((...((((((((.........................)))))))).)).((((((.....(((----------------------.((((....)))).)))..))))))...- ( -25.01) >DroSec_CAF1 21364 97 + 1 AUGCGGCUUUGAUAGCCACAUCGAUUAAUCAACGACACUUUAAUGGCUAUCUGCCGAUUUUGGACGGCA----------------------CAGUUCACGGACUUUGUAGCAAAUCAGU- .((((((...((((((((.........................)))))))).)))...........(((----------------------.((((....)))).))).))).......- ( -24.31) >DroSim_CAF1 17005 97 + 1 AUGCGGCUUUGAUAGCCACAUCGAUUAAUCAACGACACUUUAAUGGCUAUCUGCCGAUUUUGGACGGCA----------------------CAGUCCACGGACUUUGUAGCAAAUCAGU- .((((((...((((((((.........................)))))))).)))...........(((----------------------.((((....)))).))).))).......- ( -26.61) >DroEre_CAF1 25876 98 + 1 AUGCGGCUUUGAUAGCCACAUCGAUUAAUCAACGACACUUUAAUGGCUAUCUGCCGAUUUGGGACGGCA----------------------CAGUCCAUGGACUUUGUAGCAAAUCAGUC ....(((...((((((((.........................)))))))).)))((((((.....(((----------------------.((((....)))).)))..)))))).... ( -28.71) >DroYak_CAF1 25314 119 + 1 AUGCGGCUUUGAUAGCCACAUCGAUUAAUCAACGACACUUUAAUGGCUAUCUGCCGAUUUUGGACGGCACAUGGACAUGUUCCAUGGUCCACAGUCCAUGGACUUUGUAGCAAAUCAGU- .((((((...((((((((.........................)))))))).)))((((.(((((....((((((.....))))))))))).)))).............))).......- ( -34.91) >consensus AUGCGGCUUUGAUAGCCACAUCGAUUAAUCAACGACACUUUAAUGGCUAUCUGCCGAUUUUGGACGGCA______________________CAGUCCACGGACUUUGUAGCAAAUCAGU_ .(((.((...((((((((.........................))))))))(((((........))))).......................((((....))))..)).)))........ (-21.87 = -21.75 + -0.12)

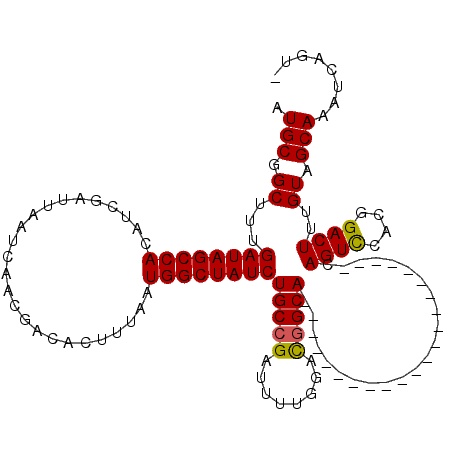
| Location | 17,964,059 – 17,964,156 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.70 |
| Mean single sequence MFE | -30.52 |
| Consensus MFE | -26.04 |
| Energy contribution | -26.64 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984660 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17964059 97 - 20766785 -ACUGAUUUGCUACAAAGUCCGUGGACUG----------------------UGCCAACCCAAAUCUGCAGAUAGCCAUUAAAGUGUCGUUGAUUAAUCGAUGUGGCUAUCAAAGCCGCAU -...((((((...((.((((....)))).----------------------))......))))))....(((((((((......((((.........))))))))))))).......... ( -24.00) >DroSec_CAF1 21364 97 - 1 -ACUGAUUUGCUACAAAGUCCGUGAACUG----------------------UGCCGUCCAAAAUCGGCAGAUAGCCAUUAAAGUGUCGUUGAUUAAUCGAUGUGGCUAUCAAAGCCGCAU -...(((((......))))).(((.....----------------------(((((........)))))(((((((((......((((.........))))))))))))).....))).. ( -27.30) >DroSim_CAF1 17005 97 - 1 -ACUGAUUUGCUACAAAGUCCGUGGACUG----------------------UGCCGUCCAAAAUCGGCAGAUAGCCAUUAAAGUGUCGUUGAUUAAUCGAUGUGGCUAUCAAAGCCGCAU -...(((((......))))).((((.((.----------------------(((((........)))))(((((((((......((((.........)))))))))))))..)))))).. ( -31.40) >DroEre_CAF1 25876 98 - 1 GACUGAUUUGCUACAAAGUCCAUGGACUG----------------------UGCCGUCCCAAAUCGGCAGAUAGCCAUUAAAGUGUCGUUGAUUAAUCGAUGUGGCUAUCAAAGCCGCAU ((.(((((.((.(((......((((.(((----------------------(((((........)))))..))))))).....))).)).))))).)).((((((((.....)))))))) ( -30.90) >DroYak_CAF1 25314 119 - 1 -ACUGAUUUGCUACAAAGUCCAUGGACUGUGGACCAUGGAACAUGUCCAUGUGCCGUCCAAAAUCGGCAGAUAGCCAUUAAAGUGUCGUUGAUUAAUCGAUGUGGCUAUCAAAGCCGCAU -...(((((.......((((....)))).(((((((((((.....))))))....))))))))))(((.(((((((((......((((.........)))))))))))))...))).... ( -39.00) >consensus _ACUGAUUUGCUACAAAGUCCGUGGACUG______________________UGCCGUCCAAAAUCGGCAGAUAGCCAUUAAAGUGUCGUUGAUUAAUCGAUGUGGCUAUCAAAGCCGCAU ........(((.....((((....)))).......................(((((........)))))(((((((((......((((.........)))))))))))))......))). (-26.04 = -26.64 + 0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:51:13 2006