
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,954,456 – 17,954,599 |
| Length | 143 |
| Max. P | 0.918242 |

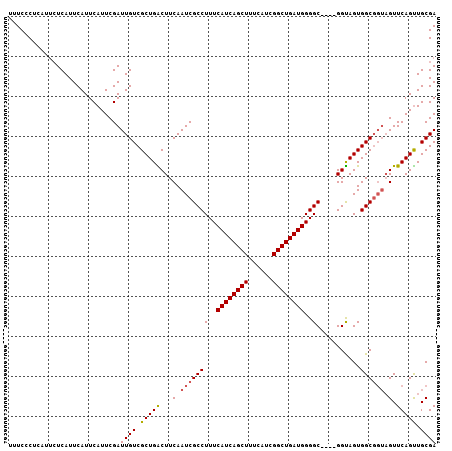
| Location | 17,954,456 – 17,954,559 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 91.11 |
| Mean single sequence MFE | -32.25 |
| Consensus MFE | -26.89 |
| Energy contribution | -27.20 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918242 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17954456 103 + 20766785 UUUCUCUCAUUCUCAUUCAUUCAUUCGAUUGUCGCUGGCUUCAAUCGCCUUUCAUCAGCUUUCAUCGGCUGAUGGGGC----GGUAGUGGCGGUAGUUCAGUUGCGA .........................((((((..(((((((.(.(((((((..((((((((......))))))))))))----))).).)))..)))).))))))... ( -31.20) >DroSec_CAF1 11723 103 + 1 UUUCCCUCAUUCUCAUUCAUUCAUUCGAUUGUCGCUGACUUCAAUCGCCUUUCAUCAGCUUUCAUCGGCUGAUGGGGC----GGUAGUGGCGGUAGUUCAGUUGCGA ......................(((.((((..((((.(((...(((((((..((((((((......))))))))))))----))))))))))..)))).)))..... ( -31.70) >DroSim_CAF1 7457 103 + 1 UUUCCCUCAUUCUCAUUCAUUCAUUCGAUUGUCGCUGACUUCAAUCGCCUUUCAUCAGCUUUCAUCGGCUGAUGGGGC----GGUAGUGGCGGUAGUUCAGUUGCGA ......................(((.((((..((((.(((...(((((((..((((((((......))))))))))))----))))))))))..)))).)))..... ( -31.70) >DroEre_CAF1 16440 97 + 1 UUUCUC------UCAUUCAUUCAUUCGAUUGUCGCUGGCUUCAAUCGCCUUUCAUCAGCUUUCAUCGGCUGAUGGGGCGGGCGGCAGUGGC----GUUCAGCUGCGA ......------..............((.((((((((.(.....((((((..((((((((......))))))))))))))..).)))))))----).))........ ( -34.40) >consensus UUUCCCUCAUUCUCAUUCAUUCAUUCGAUUGUCGCUGACUUCAAUCGCCUUUCAUCAGCUUUCAUCGGCUGAUGGGGC____GGUAGUGGCGGUAGUUCAGUUGCGA ............................((((.(((((...(.(((((((..((((((((......))))))))..)...........)))))).).))))).)))) (-26.89 = -27.20 + 0.31)


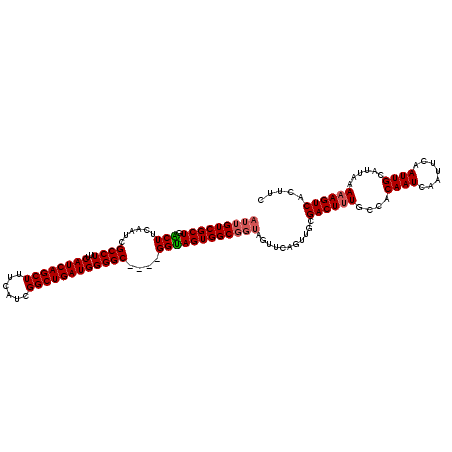
| Location | 17,954,483 – 17,954,599 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.35 |
| Mean single sequence MFE | -37.70 |
| Consensus MFE | -31.14 |
| Energy contribution | -31.70 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881732 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17954483 116 + 20766785 AUUGUCGCUGGCUUCAAUCGCCUUUCAUCAGCUUUCAUCGGCUGAUGGGGC----GGUAGUGGCGGUAGUUCAGUUGCGACUUUCCCACAAUCAAUUCAAUUGCAUUAAAAAGUCACUUC ........((((((..(((((((..((((((((......))))))))))))----))).((((.((.((((.......)))).)))))).....................)))))).... ( -34.40) >DroSec_CAF1 11750 116 + 1 AUUGUCGCUGACUUCAAUCGCCUUUCAUCAGCUUUCAUCGGCUGAUGGGGC----GGUAGUGGCGGUAGUUCAGUUGCGACUUUGCCACAAUCAAUUCAAUUGCAUUAAAAAGUCACUUC ........((((((..(((((((..((((((((......))))))))))))----))).(((((((.((((.......))))))))))).....................)))))).... ( -38.50) >DroSim_CAF1 7484 116 + 1 AUUGUCGCUGACUUCAAUCGCCUUUCAUCAGCUUUCAUCGGCUGAUGGGGC----GGUAGUGGCGGUAGUUCAGUUGCGACUUUGCCACAAUCAAUUCAAUUGCAUUAAAAAGUCACUUC ........((((((..(((((((..((((((((......))))))))))))----))).(((((((.((((.......))))))))))).....................)))))).... ( -38.50) >DroEre_CAF1 16461 116 + 1 AUUGUCGCUGGCUUCAAUCGCCUUUCAUCAGCUUUCAUCGGCUGAUGGGGCGGGCGGCAGUGGC----GUUCAGCUGCGACUGUGCCACAAUCAAUUCAAUUGCAUUAAAAAGUCACUUC ........((((((....(((((..((((((((......)))))))))))))((((.((((.((----(......))).)))))))).......................)))))).... ( -39.40) >consensus AUUGUCGCUGACUUCAAUCGCCUUUCAUCAGCUUUCAUCGGCUGAUGGGGC____GGUAGUGGCGGUAGUUCAGUUGCGACUUUGCCACAAUCAAUUCAAUUGCAUUAAAAAGUCACUUC (((((((((.(((......((((..((((((((......))))))))))))....))))))))))))...........((((((....((((.......))))......))))))..... (-31.14 = -31.70 + 0.56)

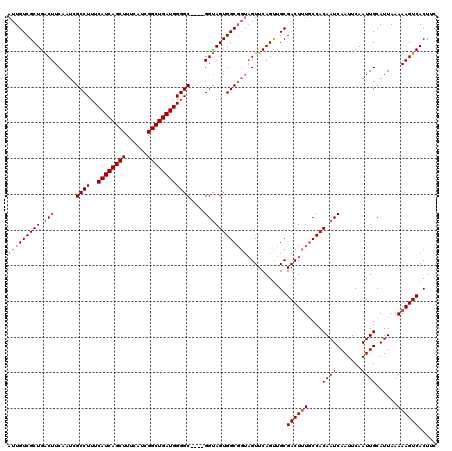
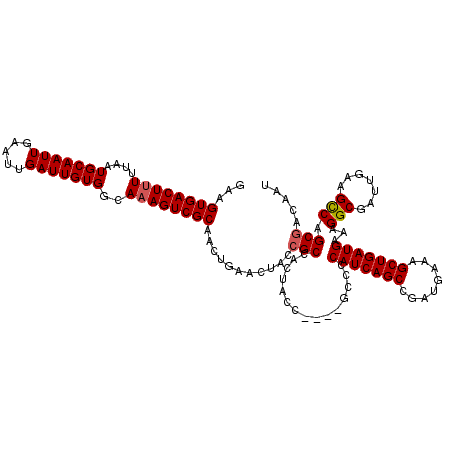
| Location | 17,954,483 – 17,954,599 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.35 |
| Mean single sequence MFE | -33.65 |
| Consensus MFE | -29.08 |
| Energy contribution | -29.32 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.614712 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17954483 116 - 20766785 GAAGUGACUUUUUAAUGCAAUUGAAUUGAUUGUGGGAAAGUCGCAACUGAACUACCGCCACUACC----GCCCCAUCAGCCGAUGAAAGCUGAUGAAAGGCGAUUGAAGCCAGCGACAAU ...((((((((((...((((((.....)))))).))))))))))....................(----((..(((((((........)))))))...(((.......))).)))..... ( -34.60) >DroSec_CAF1 11750 116 - 1 GAAGUGACUUUUUAAUGCAAUUGAAUUGAUUGUGGCAAAGUCGCAACUGAACUACCGCCACUACC----GCCCCAUCAGCCGAUGAAAGCUGAUGAAAGGCGAUUGAAGUCAGCGACAAU ...((((((((....(((((((.....)))))))..))))))))...........(((.(((.((----(((.(((((((........)))))))...))))...).)))..)))..... ( -32.90) >DroSim_CAF1 7484 116 - 1 GAAGUGACUUUUUAAUGCAAUUGAAUUGAUUGUGGCAAAGUCGCAACUGAACUACCGCCACUACC----GCCCCAUCAGCCGAUGAAAGCUGAUGAAAGGCGAUUGAAGUCAGCGACAAU ...((((((((....(((((((.....)))))))..))))))))...........(((.(((.((----(((.(((((((........)))))))...))))...).)))..)))..... ( -32.90) >DroEre_CAF1 16461 116 - 1 GAAGUGACUUUUUAAUGCAAUUGAAUUGAUUGUGGCACAGUCGCAGCUGAAC----GCCACUGCCGCCCGCCCCAUCAGCCGAUGAAAGCUGAUGAAAGGCGAUUGAAGCCAGCGACAAU ...((.((...(((((........)))))..)).))...(((((.(((...(----(((...((.....))..(((((((........)))))))...)))).....)))..)))))... ( -34.20) >consensus GAAGUGACUUUUUAAUGCAAUUGAAUUGAUUGUGGCAAAGUCGCAACUGAACUACCGCCACUACC____GCCCCAUCAGCCGAUGAAAGCUGAUGAAAGGCGAUUGAAGCCAGCGACAAU ...((((((((....(((((((.....)))))))..))))))))...........(((...............(((((((........)))))))...(((.......))).)))..... (-29.08 = -29.32 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:51:03 2006