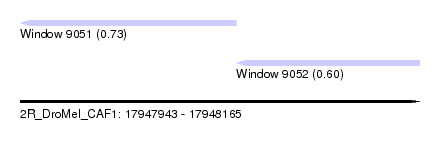
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,947,943 – 17,948,165 |
| Length | 222 |
| Max. P | 0.728977 |

| Location | 17,947,943 – 17,948,063 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.93 |
| Mean single sequence MFE | -37.36 |
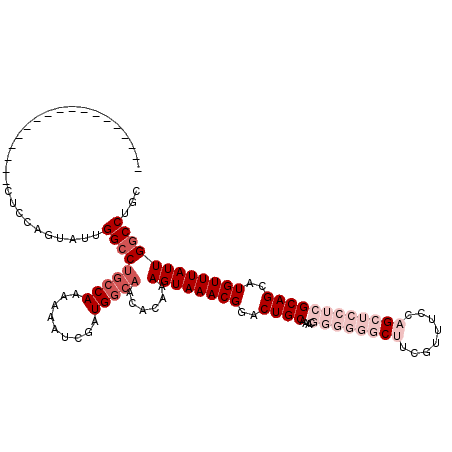
| Consensus MFE | -27.57 |
| Energy contribution | -28.38 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.728977 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17947943 120 - 20766785 AGCGGUGUUAACUUUGCCACAUUCCCCGAAGAUUUGGGCCGGGAUCCUCCCCCCAUCCCCGAAUCACCCAGUUUGCUUCUGGCUAGCUGUGGCAGACAAACUAUUGUCAGUGCAAAUAAA .(((.(........(((((((.........((((((((..(((........)))...))))))))..((((.......)))).....)))))))(((((....)))))).)))....... ( -38.10) >DroSec_CAF1 5267 120 - 1 AGCGGUGUUAACUUUGCCACGUUCCCCGAAGAUUUGGGCCGGGCUCCUCGCCCAAUCCCCGAAUCACCCAGCUUGCUUCUGGCUAGCUGUGGCAGACAAACUAUUGUCAGUGCAAAUAAA .(((.(........((((((..........((((((((..((((.....))))....))))))))....((((.((.....)).))))))))))(((((....)))))).)))....... ( -42.90) >DroSim_CAF1 1003 120 - 1 AGCGGUGUUAACUUUGCCACGUUCCCCGAAGAUUUGGGCCGGGCUCCUCCCCGAAUCCCCGCAUCGCCCAGCUUGCUUCUGGCUAGAUGUGGCAGACAAACUAUUGUCAGUGCAAAUAAA .(((.(........(((((((((.((.((((..((((((((((.((......))...))))....))))))....)))).))...)))))))))(((((....)))))).)))....... ( -39.80) >DroEre_CAF1 10049 114 - 1 AGCGGUGUUAACUUUGCCACGUUCCCCGAAGAUUUGGCCCGGGUUUCUGCC------CCCCAAUCCGCCAGUUUGCUUCUGGCUAGCUGUGGCAGACAAACUAUUGUCAGUGCAAAUAAA .(((.(........((((((..........(..((((...((((....)))------).))))..)(((((.......))))).....))))))(((((....)))))).)))....... ( -35.00) >DroYak_CAF1 10048 102 - 1 AGCGGUGUUAACUUUGCCACGUUCCCCAAAGAUUUGGGCC------------------ACGAAUCCCCCAGUUUGCUUCUGGCUAGCGGGGGCAGACAAACUAUUGUCAGUGCAAAUAAA .((((((.......)))).(((..(((((....)))))..------------------)))...(((((.(((.((.....)).))))))))..(((((....)))))...))....... ( -31.00) >consensus AGCGGUGUUAACUUUGCCACGUUCCCCGAAGAUUUGGGCCGGGCUCCUCCCC__AUCCCCGAAUCACCCAGUUUGCUUCUGGCUAGCUGUGGCAGACAAACUAUUGUCAGUGCAAAUAAA .(((.(........(((((((...((.((((..(((((.((((..............)))).....)))))....)))).)).....)))))))(((((....)))))).)))....... (-27.57 = -28.38 + 0.81)

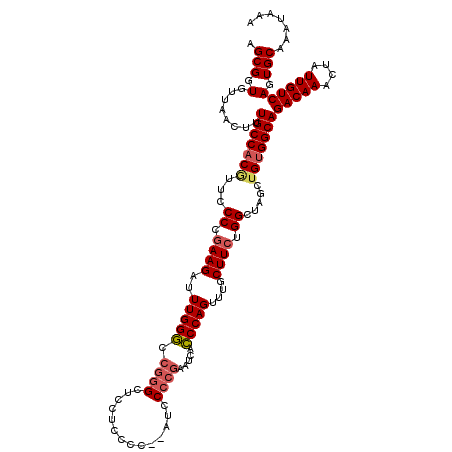
| Location | 17,948,063 – 17,948,165 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.30 |
| Mean single sequence MFE | -32.32 |
| Consensus MFE | -21.76 |
| Energy contribution | -25.36 |
| Covariance contribution | 3.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.604698 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17948063 102 - 20766785 ------------------CUUCAGUAUUGGCCUGCCAAAAAAUCGAUGGCAACACAAAGUAAACGGACUGCAAACGGGGGGCUUCGUUUCCAGCUCCUCGCAGCAUGUUUAUUGGCCUGC ------------------..........(((((((((.........)))))......((((((((..((((....((((((((........))))))))))))..))))))))))))... ( -36.10) >DroSec_CAF1 5387 101 - 1 ------------------CUCCAGUAUUGGCCUGCCAAAAAAUCGAUGGCAACACAAAGUAAACGGACUGCA-ACGGGAGGCUUCGCUUCCAGCUCCUCGCAGCAUGUUUAUUGGCCUGC ------------------..........(((((((((.........)))))......((((((((..((((.-..(((((.((........))))))).))))..))))))))))))... ( -31.90) >DroSim_CAF1 1123 101 - 1 ------------------CUCCAGUAUUGGCCUGCCAA-AAAUCGAUGGCAACACAAAGUAAACGGACUGCAAACGGGGGGCUUCGCUUCCAGCUCCUCGCAGCAUGUUUAUUGGCCUGC ------------------..........(((((((((.-.......)))))......((((((((..((((....((((((((........))))))))))))..))))))))))))... ( -36.90) >DroEre_CAF1 10163 100 - 1 ------------------CUGCAGUAUUGGCCGCCCAAAAAAUCGAUGCCAACACAAAGUAAACGGACUGCAAAAA--GCCCCCCGUUUCCAGCUCCUCGCAGCAUGUUUAUUGGCCUGC ------------------..((((..((((....)))).........(((((.(((..(.((((((...((.....--))...)))))).).(((......))).)))...))))))))) ( -22.50) >DroYak_CAF1 10150 118 - 1 ACUAUGGCAUCCAGUGUGAUGCAGUAUUGGGCUGCCAAAAAAUCGAUGGCAACACAAAGUAAACGGACUGCAAAAA--GCCCCCCGUUUGCAGCCACUCGCAGCUUGUUUAUUGGCCUGC ..(((.(((((......))))).)))...((((((((.........))))........((((((((...((.....--))...))))))))))))....((((((........)).)))) ( -34.20) >consensus __________________CUCCAGUAUUGGCCUGCCAAAAAAUCGAUGGCAACACAAAGUAAACGGACUGCAAACGGGGGGCUUCGUUUCCAGCUCCUCGCAGCAUGUUUAUUGGCCUGC ............................(((((((((.........)))))......((((((((..((((....((((((((........))))))))))))..))))))))))))... (-21.76 = -25.36 + 3.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:50:55 2006