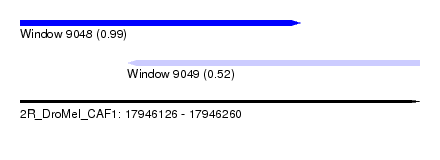
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,946,126 – 17,946,260 |
| Length | 134 |
| Max. P | 0.985526 |

| Location | 17,946,126 – 17,946,220 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 65.32 |
| Mean single sequence MFE | -39.78 |
| Consensus MFE | -13.36 |
| Energy contribution | -15.84 |
| Covariance contribution | 2.48 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.34 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985526 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17946126 94 + 20766785 UCCAGGGUGUUGCAGCUGCAAUUUCAGUUGCAUUUG------CAGUG------GCACUGGCAGUUGC--AGCUGCGGUGGGGGCU-UGCCC-----------CACCAAGUAUUUGCUCAG ....((((...((((((((((((((((((((....)------))...------..))))).))))))--))))))(((((((...-..)))-----------))))........)))).. ( -46.50) >DroVir_CAF1 4671 81 + 1 --CGAGUUGUUGGCGCUGCAAGUGCCGCAGGUGCUGCAG--------------------GUGGUUAAGGUGGUGCUGUCGGUGCAUUGGU-----------------CGUGUUUGUUCAG --(((.(.....((((((..((..((((....((((...--------------------.))))....))))..))..))))))...).)-----------------))........... ( -25.70) >DroSec_CAF1 3477 94 + 1 UCCAGGGUGUUGCAGCUGCAAUUUCAGUUGCAGUUG------CAGUG------GCACUGGCAGUUGC--AGCUGCGGUGGGUGCU-UGCCC-----------CACCAAGUAUUUGCUCAG ....((((...((((((((((((((((((((....)------))...------..))))).))))))--))))))((((((.(..-..)))-----------))))........)))).. ( -43.70) >DroEre_CAF1 8255 100 + 1 --UAGGGUGUUGCAGCUGCAAUUGCAGUUGCAGUUGCAGUUUCAGUGGCAGUGGCAGUGGCAGUGGC--AGUUGCGGUGGGUGCU-CC---------------ACCAAGUAUUUGCUCAG --..((((...((((((((.(((((..((((..((((..........))))..))))..))))).))--))))))(((((.....-))---------------)))........)))).. ( -42.90) >DroYak_CAF1 8253 111 + 1 UCCAGGGUGUUGCAGCUGCAAUUUCAAUUGCAGUUGCAGUUGCAGUG------GCAGUGGCAGUUGC--AGUAGCGAUGGGUGCU-UCCCCUCCCCCCACCGCAUCAGGUAUUUGCUCAG ....(((.(((((.(((((((((.(.(((((..((((....))))..------))))).).))))))--))).)))))(((....-..)))....)))(((......))).......... ( -40.10) >consensus UCCAGGGUGUUGCAGCUGCAAUUUCAGUUGCAGUUGCAG___CAGUG______GCACUGGCAGUUGC__AGCUGCGGUGGGUGCU_UGCCC___________CACCAAGUAUUUGCUCAG ....((((..((((((((((((....)))))))))))).....................(((((......))))).......................................)))).. (-13.36 = -15.84 + 2.48)

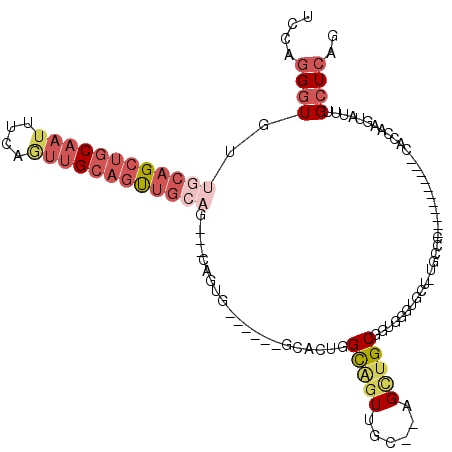
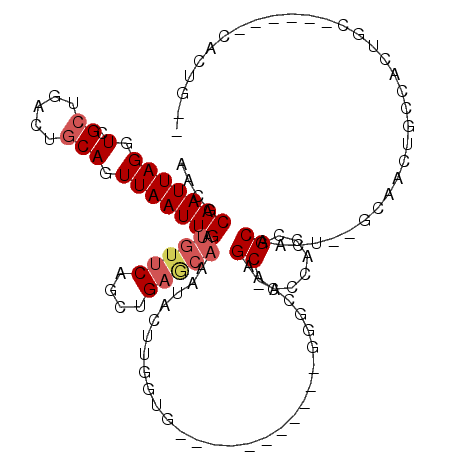
| Location | 17,946,162 – 17,946,260 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.36 |
| Mean single sequence MFE | -30.12 |
| Consensus MFE | -7.48 |
| Energy contribution | -8.36 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.25 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.523674 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17946162 98 - 20766785 AACGCAAUUAGGUCGCUGACUGCAGUUAAUUGAUGAUCAACUGAGCAAAUACUUGGUG-----------GGGCA-AGCCCCCACCGCAGCU--GCAACUGCCAGUGC------CACUG-- ..........((.(((((...((((((..(((.....)))....(((....(((((((-----------(((..-...)))))))).)).)--))))))))))))))------)....-- ( -33.10) >DroVir_CAF1 4708 83 - 1 AACACAAUUAGGUCGGC-CCUGCAGUUAAUUGACAUGCAGCUGAACAAACACG-----------------ACCAAUGCACCGACAGCACCACCUUAACCAC------------------- ..........(((((..-.((((((((....))).))))).((......))))-----------------)))..(((.......))).............------------------- ( -15.80) >DroSec_CAF1 3513 98 - 1 AACGCAAUUAGGUCGCUGACUGCAGUUAAUUGAUGUUCAGCUGAGCAAAUACUUGGUG-----------GGGCA-AGCACCCACCGCAGCU--GCAACUGCCAGUGC------CACUG-- ..........((.(((((.((((((((......(((((....))))).......((((-----------((...-....))))))..))))--)))...).))))))------)....-- ( -34.30) >DroEre_CAF1 8293 102 - 1 AACGCAAUUAGGUCGCUGACUGCAGUUAAUUGAUGUUCAGCUGAGCAAAUACUUGGU---------------GG-AGCACCCACCGCAACU--GCCACUGCCACUGCCACUGCCACUGAA ...(((....(((.((.....((((((......(((((....))))).......(((---------------((-.....)))))..))))--))....)).))).....)))....... ( -27.30) >DroYak_CAF1 8293 111 - 1 AACGCAAUUAGGUCGCUGACUGCAGUUAAUUGAUGUUCAGCUGAGCAAAUACCUGAUGCGGUGGGGGGAGGGGA-AGCACCCAUCGCUACU--GCAACUGCCACUGC------CACUGCA ...((((((((((.((((....)))).......(((((....)))))...)))))))(((((((.(((((((..-....))).))((....--))..)).)))))))------...))). ( -40.10) >consensus AACGCAAUUAGGUCGCUGACUGCAGUUAAUUGAUGUUCAGCUGAGCAAAUACUUGGUG___________GGGCA_AGCACCCACCGCAACU__GCAACUGCCACUGC______CACUG__ ....(((((((.(.((.....))).))))))).(((((....))))).............................((.......))................................. ( -7.48 = -8.36 + 0.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:50:52 2006