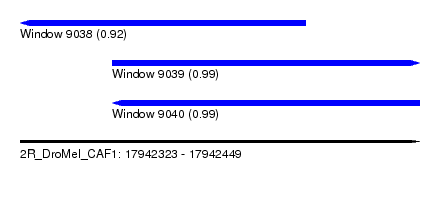
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,942,323 – 17,942,449 |
| Length | 126 |
| Max. P | 0.993253 |

| Location | 17,942,323 – 17,942,413 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.82 |
| Mean single sequence MFE | -32.10 |
| Consensus MFE | -20.62 |
| Energy contribution | -21.18 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919441 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17942323 90 - 20766785 UGAGUCAGAAGCUGACGAUCAAUUUCUGAUUACCAGUCCACUCGAAUGUGAACACCCCGUA------------------------------GCACUGGAGGGCACUGAAAUAAAGAAACA ..(((((((((.((.....)).)))))))))..((((((.(((.(.(((..((.....)).------------------------------))).).))))).))))............. ( -23.20) >DroEre_CAF1 4237 100 - 1 UGAGUCAGAAGCUGACGAUCAAUUUCUGAUUACCAGUCCACUCGAAUGUGAGCACCCCGUGGCAAU--------------------GGCGGGCAAUGGAGGGCACUGAAAUAAAGAAACA ..(((((((((.((.....)).)))))))))..((((((.(((..((....((.((((((....))--------------------)).)))).)).))))).))))............. ( -27.90) >DroYak_CAF1 3928 120 - 1 UGAGUCAGAAGCUGACGAUCAAUUUCUGAUUACCAGUCCACUCGAAUGUGAGCACUCCGUGCCACUGGAUGGCACUGGAGGCCACUGGUGAGCACUGGAGGGCACUGAAAUAAAGAAACA ..(((((((((.((.....)).)))))))))..((((.(((......))).((.(((((((((((..(.((((.(....))))))..))).)))).)))).))))))............. ( -45.20) >consensus UGAGUCAGAAGCUGACGAUCAAUUUCUGAUUACCAGUCCACUCGAAUGUGAGCACCCCGUG_CA_U____________________GG_G_GCACUGGAGGGCACUGAAAUAAAGAAACA ..(((((((((.((.....)).)))))))))..((((((.(((.(.(((..((..................................))..))).).))))).))))............. (-20.62 = -21.18 + 0.56)

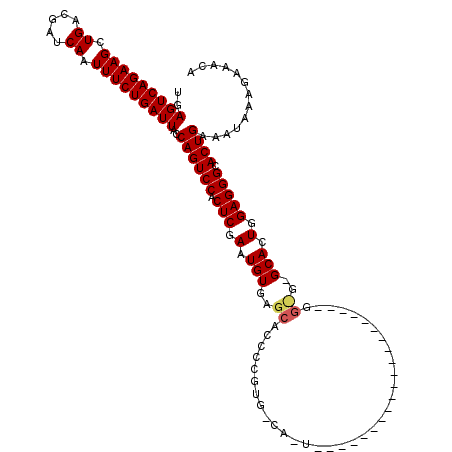
| Location | 17,942,352 – 17,942,449 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.04 |
| Mean single sequence MFE | -42.30 |
| Consensus MFE | -32.01 |
| Energy contribution | -31.90 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.81 |
| Structure conservation index | 0.76 |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.993253 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17942352 97 + 20766785 -------------------UACGGGGUGUUCACAUUCGAGUGGACUGGUAAUCAGAAAUUGAUCGUCAGCUUCUGACUCAC----UCCCACUGAUUGUGGGCACUGUGGCAAAAAUGGUU -------------------..((.((((((((((.((.(((((...(((..((((((.(((.....))).))))))...))----).))))))).)))))))))).))............ ( -34.70) >DroEre_CAF1 4271 102 + 1 --------------AUUGCCACGGGGUGCUCACAUUCGAGUGGACUGGUAAUCAGAAAUUGAUCGUCAGCUUCUGACUCAC----UCCCACUGAUUGUGGGCACUGUGGCAAAAAUGGUU --------------.(((((((..((((((((((.((.(((((...(((..((((((.(((.....))).))))))...))----).))))))).)))))))))))))))))........ ( -44.90) >DroYak_CAF1 3968 120 + 1 CUCCAGUGCCAUCCAGUGGCACGGAGUGCUCACAUUCGAGUGGACUGGUAAUCAGAAAUUGAUCGUCAGCUUCUGACUCACUCACUCGCUCUGAUUGUGGGCACUGUGGCAAAAAUGGUU ..((((((((((...)))))))(.((((((((((.((((((((...(((..((((((.(((.....))).))))))...)))...)))))).)).)))))))))).)........))).. ( -47.30) >consensus ______________A_UG_CACGGGGUGCUCACAUUCGAGUGGACUGGUAAUCAGAAAUUGAUCGUCAGCUUCUGACUCAC____UCCCACUGAUUGUGGGCACUGUGGCAAAAAUGGUU .....................((.((((((((((.((.((((((...((..((((((.(((.....))).))))))...))....)).)))))).)))))))))).))............ (-32.01 = -31.90 + -0.11)

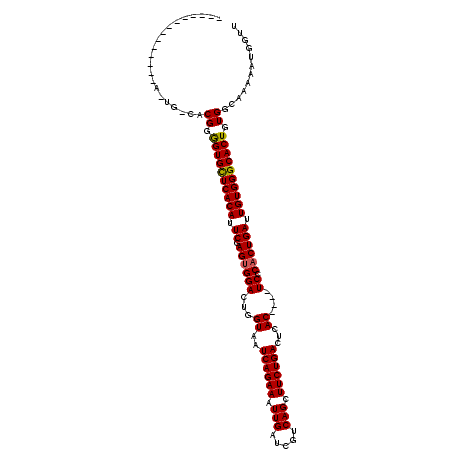
| Location | 17,942,352 – 17,942,449 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.04 |
| Mean single sequence MFE | -37.97 |
| Consensus MFE | -26.03 |
| Energy contribution | -27.37 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.52 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987348 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17942352 97 - 20766785 AACCAUUUUUGCCACAGUGCCCACAAUCAGUGGGA----GUGAGUCAGAAGCUGACGAUCAAUUUCUGAUUACCAGUCCACUCGAAUGUGAACACCCCGUA------------------- ................(((..((((.((((((((.----...(((((((((.((.....)).))))))))).....)))))).)).))))..)))......------------------- ( -28.30) >DroEre_CAF1 4271 102 - 1 AACCAUUUUUGCCACAGUGCCCACAAUCAGUGGGA----GUGAGUCAGAAGCUGACGAUCAAUUUCUGAUUACCAGUCCACUCGAAUGUGAGCACCCCGUGGCAAU-------------- ........(((((((.((((.((((.((((((((.----...(((((((((.((.....)).))))))))).....)))))).)).)))).))))...))))))).-------------- ( -42.90) >DroYak_CAF1 3968 120 - 1 AACCAUUUUUGCCACAGUGCCCACAAUCAGAGCGAGUGAGUGAGUCAGAAGCUGACGAUCAAUUUCUGAUUACCAGUCCACUCGAAUGUGAGCACUCCGUGCCACUGGAUGGCACUGGAG ..(((..........(((((.((((.((.(((.((.((.((((.(((((((.((.....)).))))))))))))).))..))))).)))).)))))..((((((.....))))))))).. ( -42.70) >consensus AACCAUUUUUGCCACAGUGCCCACAAUCAGUGGGA____GUGAGUCAGAAGCUGACGAUCAAUUUCUGAUUACCAGUCCACUCGAAUGUGAGCACCCCGUG_CA_U______________ ............(((.((((.((((.((((((((.....((((.(((((((.((.....)).)))))))))))...)))))).)).)))).))))...)))................... (-26.03 = -27.37 + 1.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:50:43 2006