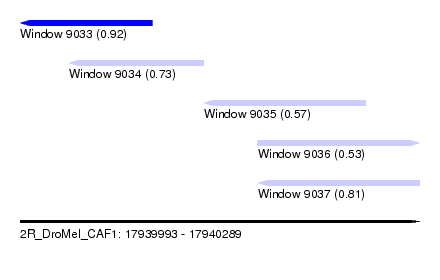
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,939,993 – 17,940,289 |
| Length | 296 |
| Max. P | 0.924854 |

| Location | 17,939,993 – 17,940,091 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.04 |
| Mean single sequence MFE | -33.80 |
| Consensus MFE | -21.01 |
| Energy contribution | -21.02 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17939993 98 - 20766785 UGCCAUUAUGGCAGCGGCAGAAGCUAGAUGCUAUU------------------GCUGCAGGGUUCCAUUAGCCGAAAUAUUAUAACAGACUUGCACGUUGCGGGGGCAAAUA----UGGC (((((((.((((.(((((((.(((.....))).))------------------))))).((...))....)))).)))...........(((((.....)))))))))....----.... ( -32.40) >DroEre_CAF1 1914 85 - 1 UGCCAUUAUGGCUGCAGCAGGAGC-------------------------------UGCAGGGUUCCAUUAGCCGAAAUAUUAUAACAGACUUGCACGUUGCGGGGGCAAAUA----UGGC ((((.(((((((((((((....))-------------------------------)))))((((.....))))......))))))....(((((.....)))))))))....----.... ( -27.50) >DroYak_CAF1 1555 120 - 1 UGCCAUUACGGCUGCAGCAGGAGCUAGUUGCUAUUGCUAUUGCUUUUGCUUUUGCUGCAGGGUUCCAUUAGCCGAAAUAUUAUAACAGACUUGCACGUUGCGGGGGCGAAUAUGUAUGGC .(((....((((((((((((((((.(((.((....))....)))...))))))))))).((...))...)))))...............(((((.....))))))))............. ( -41.50) >consensus UGCCAUUAUGGCUGCAGCAGGAGCUAG_UGCUAUU__________________GCUGCAGGGUUCCAUUAGCCGAAAUAUUAUAACAGACUUGCACGUUGCGGGGGCAAAUA____UGGC (((((((.((((.(((((...................................))))).((...))....)))).)))...........(((((.....)))))))))............ (-21.01 = -21.02 + 0.01)

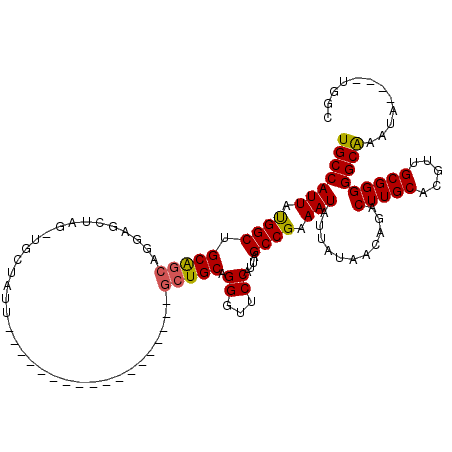
| Location | 17,940,029 – 17,940,129 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.83 |
| Mean single sequence MFE | -32.40 |
| Consensus MFE | -16.40 |
| Energy contribution | -17.18 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.730490 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17940029 100 - 20766785 AAAUAUGAGCAGUGUGAUAUGGGAUACA--CAUAGGUGAUUGCCAUUAUGGCAGCGGCAGAAGCUAGAUGCUAUU------------------GCUGCAGGGUUCCAUUAGCCGAAAUAU ...........(.((...(((((((.((--(....)))..((((.....))))(((((((.(((.....))).))------------------)))))...)))))))..)))....... ( -30.60) >DroEre_CAF1 1950 88 - 1 AAAUAUGAGCAGUGUGAUAUGGGAUACAC-CAUAGGUGAUUGCCAUUAUGGCUGCAGCAGGAGC-------------------------------UGCAGGGUUCCAUUAGCCGAAAUAU ...........(.((...(((((((...(-(((((((....)))..)))))(((((((....))-------------------------------))))).)))))))..)))....... ( -27.60) >DroYak_CAF1 1595 120 - 1 AAAUAUGAGCACUGUGAUAUGGGAUAUACACAUAGGUGAUUGCCAUUACGGCUGCAGCAGGAGCUAGUUGCUAUUGCUAUUGCUUUUGCUUUUGCUGCAGGGUUCCAUUAGCCGAAAUAU ......((((.(((((((((((......).))))(((....))).))))))(((((((((((((.(((.((....))....)))...))))))))))))).))))............... ( -39.00) >consensus AAAUAUGAGCAGUGUGAUAUGGGAUACAC_CAUAGGUGAUUGCCAUUAUGGCUGCAGCAGGAGCUAG_UGCUAUU__________________GCUGCAGGGUUCCAUUAGCCGAAAUAU .............((...(((((((.........(((....))).......(((((((...................................))))))).)))))))..))........ (-16.40 = -17.18 + 0.78)

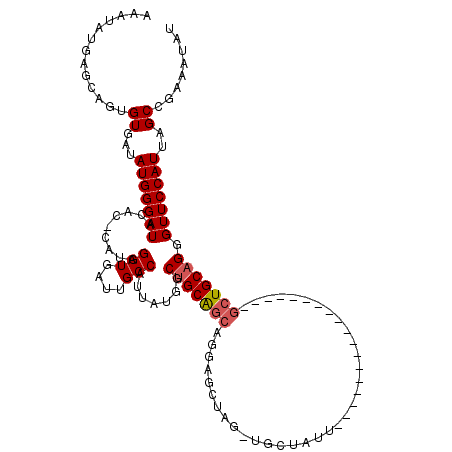
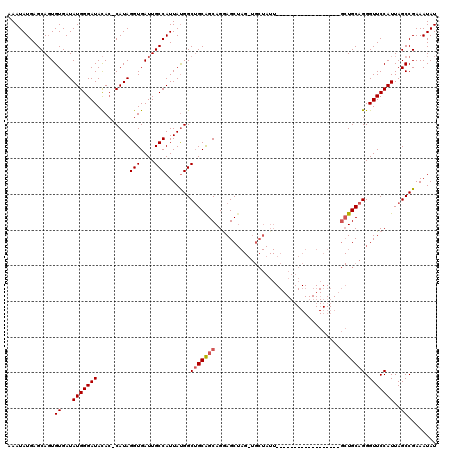
| Location | 17,940,129 – 17,940,249 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -41.00 |
| Consensus MFE | -39.04 |
| Energy contribution | -39.37 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.569112 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17940129 120 - 20766785 CUGUGGCGCUGCAUGUGUAAAAUUACCCAAUUCCAAAUGCAUUUCGGUGCAUUAGGCAGGUGGGGCACUGGAAUUUGCAUAUUUGCAACGAGUUUUCAGCUCCGCCGCCGGCAGAACCUC ....((..((((.((.(((....))).))......(((((((....))))))).(((.((((((((..(((((.(((((....))))).....)))))))))))))))).))))..)).. ( -42.30) >DroEre_CAF1 2038 120 - 1 CUGUGGCGCUGCAUGUGUAAAAUUACCCAAUUCCAAAUGCAUUUCGGUGCAUUAGGCAGGUGGGGCACUGGAAUUUGCAUAUUUGCAACGAGUUUUCAGCUCCGCCGCCGGCAGACCGUC ....((((((((.((.(((....))).))......(((((((....))))))).(((.((((((((..(((((.(((((....))))).....)))))))))))))))).))))..)))) ( -41.30) >DroYak_CAF1 1715 120 - 1 CUGUGGCGCUGCAUGUGUAAAAUUACCGAAUUCCAAAUGCAUUUCGGUGCAUUAGGCAGGUGGGGCACUGGAAUUUGCAUAUUUGCAACGAGUUUUCACCUCCGCCGCCGGCAGACCGUC ((((((((..((...........(((((((............))))))).....((.(((((..(.(((.(...(((((....)))))).))))..))))))))))))).))))...... ( -39.40) >consensus CUGUGGCGCUGCAUGUGUAAAAUUACCCAAUUCCAAAUGCAUUUCGGUGCAUUAGGCAGGUGGGGCACUGGAAUUUGCAUAUUUGCAACGAGUUUUCAGCUCCGCCGCCGGCAGACCGUC ((((.((((((...(((((..................)))))..))))))....(((.((((((((..(((((.(((((....))))).....)))))))))))))))).))))...... (-39.04 = -39.37 + 0.33)

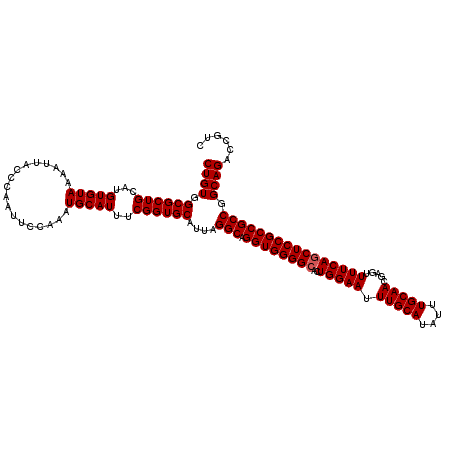
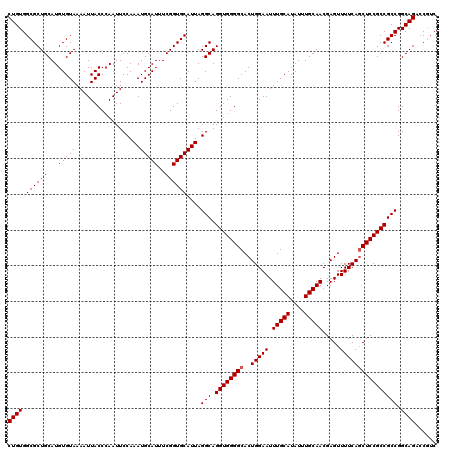
| Location | 17,940,169 – 17,940,289 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -32.37 |
| Consensus MFE | -30.33 |
| Energy contribution | -31.33 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.94 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.529290 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17940169 120 + 20766785 AUGCAAAUUCCAGUGCCCCACCUGCCUAAUGCACCGAAAUGCAUUUGGAAUUGGGUAAUUUUACACAUGCAGCGCCACAGGCGACAUUCUGGGCUUAAAAAUGCAUUUUGGCUUUUGCAU (((((((..(((((((((......((.((((((......)))))).))....))))).........(((((..(((.((((......))))))).......))))).))))..))))))) ( -36.40) >DroEre_CAF1 2078 120 + 1 AUGCAAAUUCCAGUGCCCCACCUGCCUAAUGCACCGAAAUGCAUUUGGAAUUGGGUAAUUUUACACAUGCAGCGCCACAGGCGACAUUCUGGGCUUAAAAAUGCAUUUUGGCUUUUGGAU ........(((((.(((..((((.((.((((((......)))))).))....))))..........(((((..(((.((((......))))))).......)))))...)))..))))). ( -32.10) >DroYak_CAF1 1755 120 + 1 AUGCAAAUUCCAGUGCCCCACCUGCCUAAUGCACCGAAAUGCAUUUGGAAUUCGGUAAUUUUACACAUGCAGCGCCACAGGCGACAUUCUGGGCUUAAAAAUGCAUUUUGGCUUUUCCAU .((((.....(((........))).....))))(((((((((((((........(((....))).........(((.((((......)))))))....)))))))))))))......... ( -28.60) >consensus AUGCAAAUUCCAGUGCCCCACCUGCCUAAUGCACCGAAAUGCAUUUGGAAUUGGGUAAUUUUACACAUGCAGCGCCACAGGCGACAUUCUGGGCUUAAAAAUGCAUUUUGGCUUUUGCAU (((((((..(((((((((......((.((((((......)))))).))....))))).........(((((..(((.((((......))))))).......))))).))))..))))))) (-30.33 = -31.33 + 1.00)

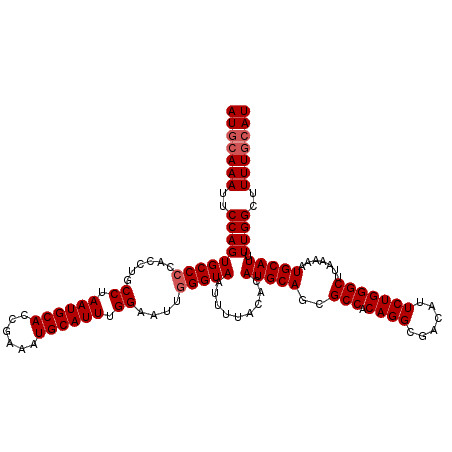
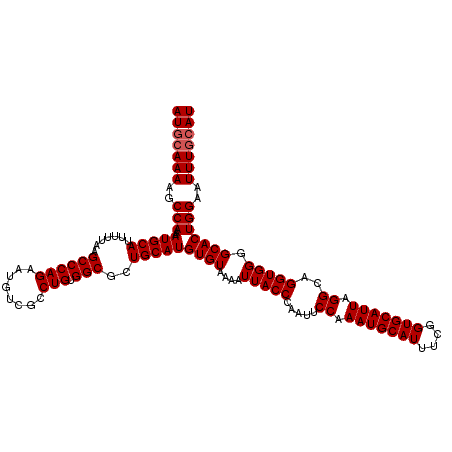
| Location | 17,940,169 – 17,940,289 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -36.74 |
| Consensus MFE | -34.53 |
| Energy contribution | -35.20 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.806447 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17940169 120 - 20766785 AUGCAAAAGCCAAAAUGCAUUUUUAAGCCCAGAAUGUCGCCUGUGGCGCUGCAUGUGUAAAAUUACCCAAUUCCAAAUGCAUUUCGGUGCAUUAGGCAGGUGGGGCACUGGAAUUUGCAU (((((((..(((..(((((.......((((((........))).)))..)))))((((.......((((...((.(((((((....))))))).))....)))))))))))..))))))) ( -39.01) >DroEre_CAF1 2078 120 - 1 AUCCAAAAGCCAAAAUGCAUUUUUAAGCCCAGAAUGUCGCCUGUGGCGCUGCAUGUGUAAAAUUACCCAAUUCCAAAUGCAUUUCGGUGCAUUAGGCAGGUGGGGCACUGGAAUUUGCAU .((((...(((....(((((......((((((........))).))).......))))).......(((...((.(((((((....))))))).))....))))))..))))........ ( -33.82) >DroYak_CAF1 1755 120 - 1 AUGGAAAAGCCAAAAUGCAUUUUUAAGCCCAGAAUGUCGCCUGUGGCGCUGCAUGUGUAAAAUUACCGAAUUCCAAAUGCAUUUCGGUGCAUUAGGCAGGUGGGGCACUGGAAUUUGCAU .(((.....)))..(((((..(((..((((((........))).)))(((.(((.(((..((((((((((............))))))).)))..))).))).)))....)))..))))) ( -37.40) >consensus AUGCAAAAGCCAAAAUGCAUUUUUAAGCCCAGAAUGUCGCCUGUGGCGCUGCAUGUGUAAAAUUACCCAAUUCCAAAUGCAUUUCGGUGCAUUAGGCAGGUGGGGCACUGGAAUUUGCAU (((((((..(((..(((((.......((((((........))).)))..)))))((((....(((((.....((.(((((((....))))))).))..))))).)))))))..))))))) (-34.53 = -35.20 + 0.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:50:41 2006