
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,883,853 – 17,883,952 |
| Length | 99 |
| Max. P | 0.852547 |

| Location | 17,883,853 – 17,883,952 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 72.19 |
| Mean single sequence MFE | -24.39 |
| Consensus MFE | -14.32 |
| Energy contribution | -14.52 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.852547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
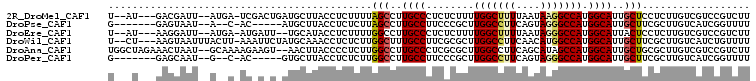
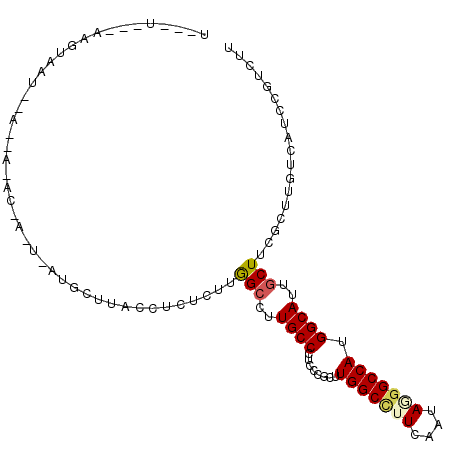
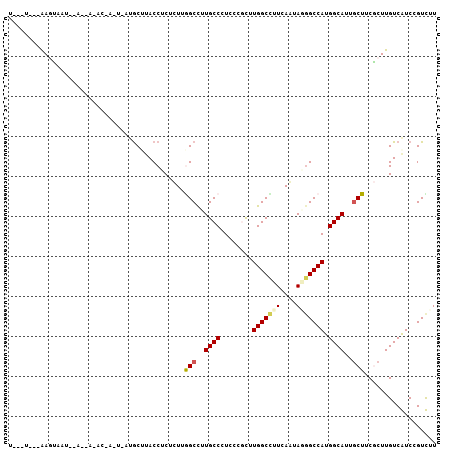
>2R_DroMel_CAF1 17883853 99 - 20766785 U--AU---GACGAUU--AUGA-UCGACUGAUGCUUACCUCUUUUAGCCUUGCCCUCUCUUUUGGCUUUUAAUAAGGCCAUGGCAUUGCUCCUCUUGUCGUCCGUCUU .--..---((((((.--..((-..((..((((((..(((..((.((((..............))))...))..)))....))))))..)).))..))))))...... ( -23.54) >DroPse_CAF1 26839 90 - 1 G-------GAGUAAU--A--C-AC-----AUGCUUACCUCUCUUAGCCUUGCCUUCCCGCUUGGCCUUCAGUAGGGCCAUGGCAUUGCUUCGCUUGUCAUCGGUUUU (-------(((((((--.--(-.(-----(((....(((((....(((..((......))..)))....)).)))..))))).))))))))................ ( -23.00) >DroEre_CAF1 1085 97 - 1 U--AU---AAGGAUU--AUGA-AUGAUU--UGCAUACCUCUUUUGGCCUUGCCCUCUCUUUUGGCUUUUAAUAGGGCCAUGGCAUUACUCCUCUUGUCGUCCGUCUU .--..---..((((.--....-......--.............((((((((((.........))).......))))))).((((..........))))))))..... ( -17.81) >DroWil_CAF1 1196 101 - 1 U--CU---AAGUAAUUUACUU-AAAUUCUAUGCAAACCUCUCUUGGCUUUGCCUUCGCGCUUGGCCUUCAACAUGGCCAUGGCAUUGCUUCGCUUGUCAUCUGUUUU .--..---(((((((......-.........(((((((......)).)))))......((((((((........))))).))))))))))................. ( -22.80) >DroAna_CAF1 29640 103 - 1 UGGCUAGAAACUAAU--GCAAAAGAAGU--AACUUACCCCUCUUGGCCUUGCCCUCGCGCUUGGCCUUCAGCAUAGCCAUGGCAUUGCUGCGCUUGUCGUCCGUCUU .(((((........(--((..((((.((--(...)))...))))((((..((......))..))))....))))))))((((((..((...)).))))))....... ( -23.70) >DroPer_CAF1 26789 90 - 1 G-------GAGCAAU--G--C-AC-----GUGCUUACCUCUCUUGGCCUUGCCUUCCCGCUUGGCCUUCAGUAGGGCCAUGGCAUUGCUUCGCUUGUCAUCGGUUUU (-------(((((((--(--(-.(-----(((....(((((...((((..((......))..))))...)).)))..))))))))))))))................ ( -35.50) >consensus U___U___AAGUAAU__A__A_AC_A_U_AUGCUUACCUCUCUUGGCCUUGCCCUCCCGCUUGGCCUUCAAUAGGGCCAUGGCAUUGCUUCGCUUGUCAUCCGUCUU ............................................(((..((((........(((((((....))))))).))))..))).................. (-14.32 = -14.52 + 0.19)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:50:14 2006