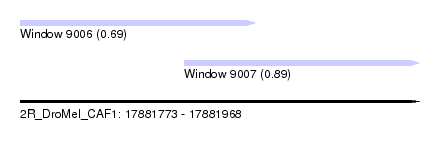
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,881,773 – 17,881,968 |
| Length | 195 |
| Max. P | 0.891020 |

| Location | 17,881,773 – 17,881,888 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.75 |
| Mean single sequence MFE | -38.20 |
| Consensus MFE | -29.40 |
| Energy contribution | -29.84 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.689712 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17881773 115 + 20766785 ACCCACUUACCAGCGUGCAGACAAAUGUUUGCCUAUUACAGGGUACACAAAGUCAGCAUUCCCGCCCAGCCAUUCGGUAUCAUCAUGCAAAUUGAAGGGGG----CUGG-GAUAGGGAUA ..............((((.(((...(((.(((((......))))).)))..))).))))((((.(((((((.((((((............))))))...))----))))-)...)))).. ( -39.40) >DroSec_CAF1 46741 115 + 1 ACCCACUUACCAGCGUGCAGACAGAUGUUUGCCUAUUACAGGGUACACAAAGUCAGCAUUCCCGCCCAGCCAUUCGGUAUCAUCAUGCAAAUUGAAGGGGG----CUGG-GAUAGGGAUA ..............((((.(((...(((.(((((......))))).)))..))).))))((((.(((((((.((((((............))))))...))----))))-)...)))).. ( -39.40) >DroSim_CAF1 47104 115 + 1 ACCCACUUACCAGCGUGCAGACAAAUGUUUGCCUAUUACAGGGUACACAAAGUCAGCAUUCCCGCCCAGCCAUUCGGUAUCAUCAUGCAAAUUGAAGGGGG----CUGG-GAUAGGGAUA ..............((((.(((...(((.(((((......))))).)))..))).))))((((.(((((((.((((((............))))))...))----))))-)...)))).. ( -39.40) >DroEre_CAF1 49175 119 + 1 ACCCACUUACCACCGUGCAGAUAAAUGUUUGCCUAUUACAGGGUACACAAAGUCAGCAUUCCCGCCCAGCCAUUCGCUAUCAUCAUGCAAAUUGAAGGGGGGGUGCUGG-GAUAGGGAUA ..............((((.(((...(((.(((((......))))).)))..))).))))((((.((((((.((((.((....(((.......)))..)).)))))))))-)...)))).. ( -37.30) >DroYak_CAF1 46418 116 + 1 ACCCACUUACCAGCGUGCAGACAAAUGUUUGCCUAUUACAGGGUACACAAAGUCAGCAUUCCCGCCCAGCCAUUCGCUAUCAUCAUGCAAAUUGAAGGGGG----CUGGAGAUAGGGAUA .(((..((.(((((((((.(((...(((.(((((......))))).)))..))).))))((((....(((.....)))....(((.......))).)))))----)))).))..)))... ( -35.50) >consensus ACCCACUUACCAGCGUGCAGACAAAUGUUUGCCUAUUACAGGGUACACAAAGUCAGCAUUCCCGCCCAGCCAUUCGGUAUCAUCAUGCAAAUUGAAGGGGG____CUGG_GAUAGGGAUA .(((..((.((((.((((.(((...(((.(((((......))))).)))..))).))))..((.(((.(((....)))....(((.......))).)))))....)))).))..)))... (-29.40 = -29.84 + 0.44)

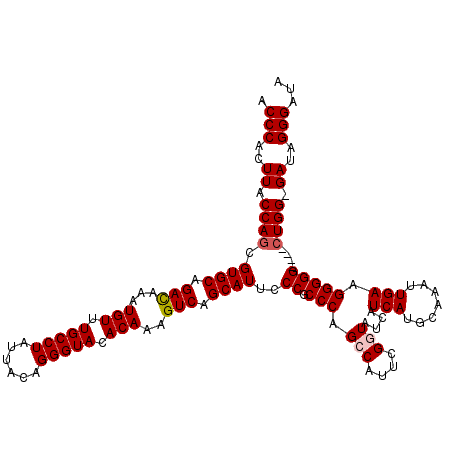
| Location | 17,881,853 – 17,881,968 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.82 |
| Mean single sequence MFE | -43.22 |
| Consensus MFE | -30.08 |
| Energy contribution | -31.36 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.33 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.891020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17881853 115 + 20766785 CAUCAUGCAAAUUGAAGGGGG----CUGG-GAUAGGGAUAAUGGGGAUGCCGAAAUUUGCAUUUCAAUUCUGCCGCCGCCGGCAUUUCCAUUCCCCUGCACCUCCACCCACGUCCCCACC ..(((.......))).(((((----((((-(.(((((..(((((..((((((......(((.........)))......))))))..))))).)))))........)))).))))))... ( -43.60) >DroSec_CAF1 46821 115 + 1 CAUCAUGCAAAUUGAAGGGGG----CUGG-GAUAGGGAUAAUGGGGAUGCCGAAAUUUGCAUUUCAAUUCUGCCGUCGCCGGCAUUUCCAUUCCCCUGCACCUCCACUCACGUCCCCACC ..(((.......))).(((((----(.((-..(((((..(((((..((((((......(((.........)))......))))))..))))).)))))..)).........))))))... ( -41.30) >DroSim_CAF1 47184 115 + 1 CAUCAUGCAAAUUGAAGGGGG----CUGG-GAUAGGGAUAAUGGGGAUGCCGAAAUUUGCAUUUCAAUUCUGCCGUCGCCGGCAUUUCCAUUCCCCUGCACCUCCACCCACGUCCCCACC ..(((.......))).(((((----((((-(.(((((..(((((..((((((......(((.........)))......))))))..))))).)))))........)))).))))))... ( -43.60) >DroEre_CAF1 49255 113 + 1 CAUCAUGCAAAUUGAAGGGGGGGUGCUGG-GAUAGGGAUAAUGGGGAUGCCAAAAUUUGCAUUUAAAUUCUGCCGUCGCCGGCAUUUCCAUUCCCCUGCUCCUCCACCC-----CCC-AC ..(((.......)))..((((((((..((-..(((((..(((((..(((((.......(((.........))).......)))))..))))).)))))..))..)))))-----)))-.. ( -50.74) >DroYak_CAF1 46498 111 + 1 CAUCAUGCAAAUUGAAGGGGG----CUGGAGAUAGGGAUAAUGGGGAUGCCAAAAUUUGCAUUUCAAUUCUGCCGUCGCCGGCAUUUCCAUUCCUCCGCUCUUCCGCAC-----CCGAAC ..(((.......))).(((.(----(.(((((..(((..(((((..(((((.......(((.........))).......)))))..)))))..)))..))))).)).)-----)).... ( -36.84) >consensus CAUCAUGCAAAUUGAAGGGGG____CUGG_GAUAGGGAUAAUGGGGAUGCCGAAAUUUGCAUUUCAAUUCUGCCGUCGCCGGCAUUUCCAUUCCCCUGCACCUCCACCCACGUCCCCACC ..(((.......))).(((((.....(((...(((((..(((((..((((((......(((.........)))......))))))..))))).))))).....)))......)))))... (-30.08 = -31.36 + 1.28)

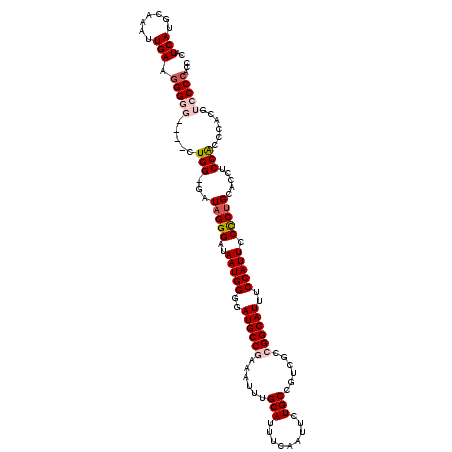
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:50:12 2006