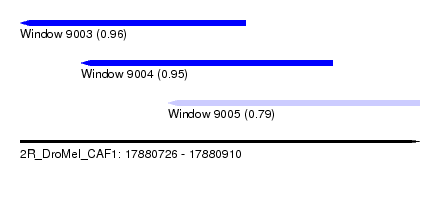
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,880,726 – 17,880,910 |
| Length | 184 |
| Max. P | 0.956053 |

| Location | 17,880,726 – 17,880,830 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.06 |
| Mean single sequence MFE | -30.02 |
| Consensus MFE | -15.08 |
| Energy contribution | -15.33 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.956053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
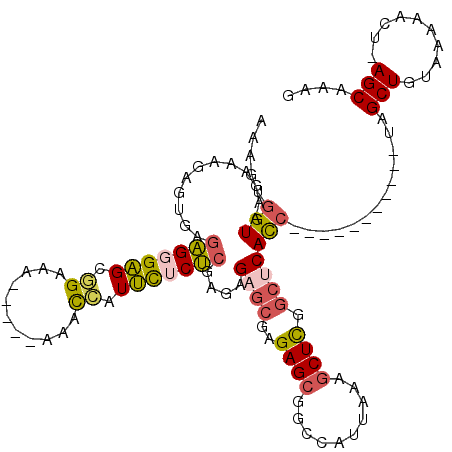
>2R_DroMel_CAF1 17880726 104 - 20766785 AAAGUGGUAAGAAAGAGUGAGAGGGAGCGGAAA----AAAACCAUUCUCUCGAGAGAGCGAGAGCGGCCAUUAAAGCUCGGCUCAUC-----------UAGCUGUUAAAACU-AGCAAAG ..((..((.(((........(((((((.((...----....)).)))))))....((((..((((..........)))).)))).))-----------).))..))......-....... ( -28.50) >DroSec_CAF1 45703 103 - 1 AAAGUGGUAAGAAAGAGUGAGAGGGAGUGGAAA-----ACACCAUUCUCUCGAGAGAGCGAGAGCGGCCAUUAAAGCUCGGCUCACC-----------UAGCUGUAAAACCU-AGCAAAG .....(((............((((((((((...-----...))))))))))....((((..((((..........)))).)))))))-----------..((((.......)-))).... ( -33.50) >DroSim_CAF1 46066 103 - 1 AAAGUGGUAAGAAAGAGUGAGAGGGAGUGGAAG-----AAACCAUUUUCUCGAGAGAGCGAGAGCGGCCAUUAAAGCUCGGCUCACC-----------UAGCUGUAAAAACU-AGCAAAG .....(((............(((..(((((...-----...)))))..)))....((((..((((..........)))).)))))))-----------..((((.......)-))).... ( -31.00) >DroEre_CAF1 48084 108 - 1 AAAGUGGUAAGAAAGAGUGAGAGGGAGCGGCAAAGAAAAAACAAUUCACUCGCGAGCGCGAGAGCGGCCAUUAAAGCUCGGCUCACC-----------UAGCUGUAAAAACU-AGCAAAG .....(((.((...((((......(.((.((...(((.......))).(((((....))))).)).)))......))))..)).)))-----------..((((.......)-))).... ( -28.50) >DroWil_CAF1 63155 103 - 1 AC----GUAGAGAC------GGGGGUGUUGUUU-----GAGUGAUGUGCCAGAGAGAGCGAGAGAGUG-AG-CGAGCUUUUGCCACCUUUUGUUGCUGUCGCUGUAACAACUAAGCAAAG .(----((....))------)......((((((-----.(((..(((..(((.((.(((((.((((((-.(-(((....)))))))..))).))))).)).)))..)))))))))))).. ( -28.70) >DroYak_CAF1 45351 105 - 1 AAAGUGGUAAGAAAGAGUGAGAGGGAGCGGAAGAGA---AACCCUUCUCUCGAGAGAGCGAGAGCGGCCAGUAAAGCUCGGCUCACC-----------UAGCUGUAAAAACU-AGCAAAG .....(((......(((.((.((((..(....)...---..)))))).)))....((((..((((..........)))).)))))))-----------..((((.......)-))).... ( -29.90) >consensus AAAGUGGUAAGAAAGAGUGAGAGGGAGCGGAAA_____AAACCAUUCUCUCGAGAGAGCGAGAGCGGCCAUUAAAGCUCGGCUCACC___________UAGCUGUAAAAACU_AGCAAAG .....(((............(((((((.((...........)).)))))))....((((..((((..........)))).))))))).............(((..........))).... (-15.08 = -15.33 + 0.26)

| Location | 17,880,754 – 17,880,870 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.11 |
| Mean single sequence MFE | -39.83 |
| Consensus MFE | -30.11 |
| Energy contribution | -30.12 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.945245 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17880754 116 - 20766785 GCGUUGGCGUUGCGUUUUUGUUGCGCCGGCUGUUUUGCCAAAAGUGGUAAGAAAGAGUGAGAGGGAGCGGAAA----AAAACCAUUCUCUCGAGAGAGCGAGAGCGGCCAUUAAAGCUCG ..((((((((.(((....))).))))))))..((((((((....))))))))..((((..(((((((.((...----....)).)))))))......((....))..........)))). ( -41.20) >DroSec_CAF1 45731 115 - 1 GCGUUGGCGUUGCGUUUUUGUUGCGCCGGCUGUUUUGCCAAAAGUGGUAAGAAAGAGUGAGAGGGAGUGGAAA-----ACACCAUUCUCUCGAGAGAGCGAGAGCGGCCAUUAAAGCUCG ..((((((((.(((....))).))))))))..((((((((....))))))))..((((..((((((((((...-----...))))))))))......((....))..........)))). ( -46.30) >DroSim_CAF1 46094 115 - 1 GCGUUGGCGUUGCGUUUUUGUUGCGCCGGCUGUUUUGCCAAAAGUGGUAAGAAAGAGUGAGAGGGAGUGGAAG-----AAACCAUUUUCUCGAGAGAGCGAGAGCGGCCAUUAAAGCUCG ..((((((((.(((....))).))))))))..((((((((....))))))))..((((..(((..(((((...-----...)))))..)))......((....))..........)))). ( -43.80) >DroEre_CAF1 48112 120 - 1 GCGUUGGCGUUGCGUUUUUGUUGCGCCGGCUGUUUUGCCAAAAGUGGUAAGAAAGAGUGAGAGGGAGCGGCAAAGAAAAAACAAUUCACUCGCGAGCGCGAGAGCGGCCAUUAAAGCUCG ..((((((((.(((....))).))))))))..((((((((....))))))))..((((......(.((.((...(((.......))).(((((....))))).)).)))......)))). ( -42.70) >DroWil_CAF1 63195 99 - 1 GCGUUGGCGUUGGAC----AUUGCGCCGGUGAAUUUAGCGAC----GUAGAGAC------GGGGGUGUUGUUU-----GAGUGAUGUGCCAGAGAGAGCGAGAGAGUG-AG-CGAGCUUU .((..(((((.....----...)))))..))......((..(----((....))------)((.(..(..(..-----..)..)..).)).......))...(((((.-..-...))))) ( -22.50) >DroYak_CAF1 45379 117 - 1 GCGUUGGCGUUGCGUUUUUGUUGCGCCGGCUGUUUUGCCAAAAGUGGUAAGAAAGAGUGAGAGGGAGCGGAAGAGA---AACCCUUCUCUCGAGAGAGCGAGAGCGGCCAGUAAAGCUCG ((...(((((.(((....))).)))))(((((((((((((....))))))))........(((((..(....)...---..)))))((((((......)))))))))))......))... ( -42.50) >consensus GCGUUGGCGUUGCGUUUUUGUUGCGCCGGCUGUUUUGCCAAAAGUGGUAAGAAAGAGUGAGAGGGAGCGGAAA_____AAACCAUUCUCUCGAGAGAGCGAGAGCGGCCAUUAAAGCUCG ((((((((((.(((....))).))))))))..((((((((....))))))))........(((((((.((...........)).)))))))......))..((((..........)))). (-30.11 = -30.12 + 0.01)


| Location | 17,880,794 – 17,880,910 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.16 |
| Mean single sequence MFE | -39.06 |
| Consensus MFE | -30.33 |
| Energy contribution | -31.67 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.785779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
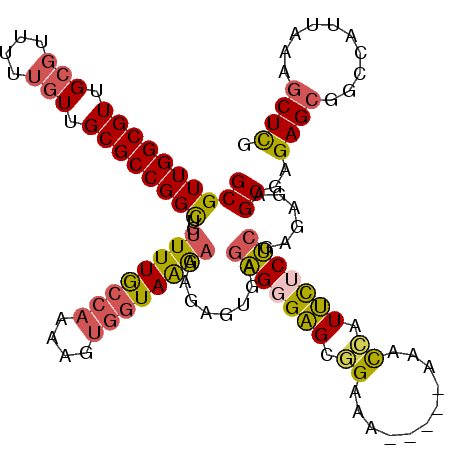
>2R_DroMel_CAF1 17880794 116 - 20766785 GCCAUUUUUGGGUUGAAAGCUCUUGUCCGGCGGCGCUGUUGCGUUGGCGUUGCGUUUUUGUUGCGCCGGCUGUUUUGCCAAAAGUGGUAAGAAAGAGUGAGAGGGAGCGGAAA----AAA .(((....))).......((((((.(((((((((((....)))))(((((.(((....))).))))).))))((((((((....))))))))......))..)))))).....----... ( -39.60) >DroSec_CAF1 45771 115 - 1 GCCAUUUUUGGGUUGAAAGCUCUUGUCCGGCGGCGCUGUUGCGUUGGCGUUGCGUUUUUGUUGCGCCGGCUGUUUUGCCAAAAGUGGUAAGAAAGAGUGAGAGGGAGUGGAAA-----AC .((((((((.........((((((...(((((((((....)))))(((((.(((....))).))))).))))((((((((....))))))))))))))....))))))))...-----.. ( -40.92) >DroSim_CAF1 46134 115 - 1 GCCAUUUUUGGGUUGAAAGCUCUUGUCCGGCGGCGCUGUUGCGUUGGCGUUGCGUUUUUGUUGCGCCGGCUGUUUUGCCAAAAGUGGUAAGAAAGAGUGAGAGGGAGUGGAAG-----AA .((((((((.........((((((...(((((((((....)))))(((((.(((....))).))))).))))((((((((....))))))))))))))....))))))))...-----.. ( -40.92) >DroEre_CAF1 48152 120 - 1 GCCAUUUUUGGGUUGAAAGCUCUUGUCCGGCGGCGCUGUUGCGUUGGCGUUGCGUUUUUGUUGCGCCGGCUGUUUUGCCAAAAGUGGUAAGAAAGAGUGAGAGGGAGCGGCAAAGAAAAA (((.((((......))))((((((.(((((((((((....)))))(((((.(((....))).))))).))))((((((((....))))))))......))..)))))))))......... ( -42.10) >DroWil_CAF1 63233 93 - 1 GCCAUUUUUGGGUGGAAACUU--------ACGGCUCUGUUGCGUUGGCGUUGGAC----AUUGCGCCGGUGAAUUUAGCGAC----GUAGAGAC------GGGGGUGUUGUUU-----GA .(((((....)))))((((..--------(((.(((((((((((((((((.....----...))))).((.......)))))----))...)))------)))).))).))))-----.. ( -29.10) >DroYak_CAF1 45419 117 - 1 GCCAUUUUUGGGUUGAAAGCUCUUGUCCGGCGGCGCUGUUGCGUUGGCGUUGCGUUUUUGUUGCGCCGGCUGUUUUGCCAAAAGUGGUAAGAAAGAGUGAGAGGGAGCGGAAGAGA---A .(((....))).........((((.((((.(..(.((.(..(((((((((.(((....))).))))))))..((((((((....))))))))....)..).)).).))))).))))---. ( -41.70) >consensus GCCAUUUUUGGGUUGAAAGCUCUUGUCCGGCGGCGCUGUUGCGUUGGCGUUGCGUUUUUGUUGCGCCGGCUGUUUUGCCAAAAGUGGUAAGAAAGAGUGAGAGGGAGCGGAAA_____AA .(((....))).......((((((...(((((((((....)))))(((((.(((....))).))))).))))((((((((....))))))))..........))))))............ (-30.33 = -31.67 + 1.34)

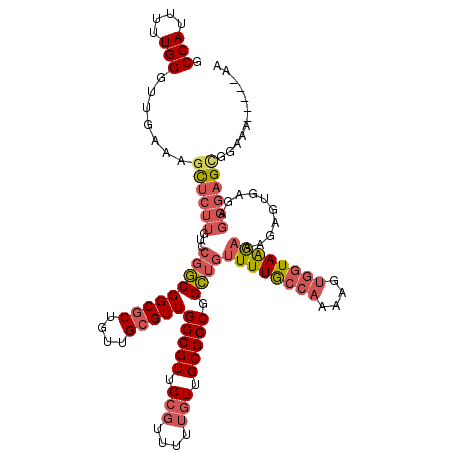
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:50:10 2006