
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,879,534 – 17,879,718 |
| Length | 184 |
| Max. P | 0.999856 |

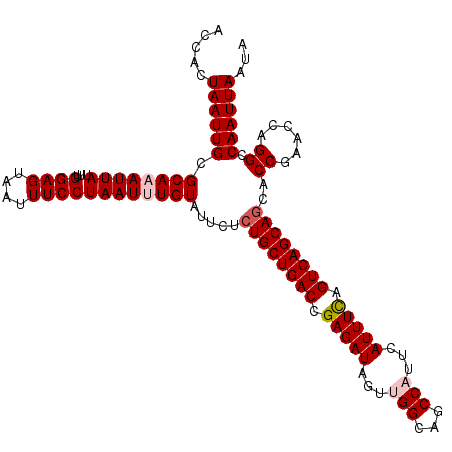
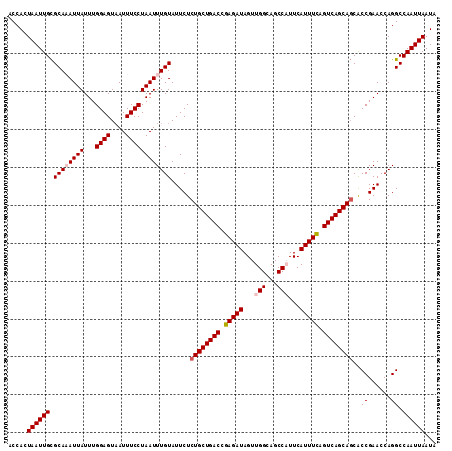
| Location | 17,879,534 – 17,879,647 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 97.17 |
| Mean single sequence MFE | -29.52 |
| Consensus MFE | -27.10 |
| Energy contribution | -27.94 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.70 |
| SVM RNA-class probability | 0.999540 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17879534 113 + 20766785 ACCACUAAUUGUGCAAAUUAUUUGGAGUAAUUUCCUAAUUUGUAUUCUCUGCUGACCGAGAUAGUUGGCAGCCAUUCAUUUCAGUCAGCACCACCGAACCAGGCCAAUUAAUA .....(((((((((((((((...((((....))))))))))))).....(((((((.(((((...(((...)))...))))).)))))))...((......)).))))))... ( -29.10) >DroSec_CAF1 44443 113 + 1 ACCACUAAUUGCGCAAAUUAUUUGGAGUAAUUUCCUAAUUUGUAUUCUCUGCUGACCGAGAUAGUUGGCAGCCAUUCAUUUCAGUCAGCAGCGCCGAACCAGGCCAAUUAAUA .....((((((.((((((((...((((....)))))))))))).....((((((((.(((((...(((...)))...))))).)))))))).(((......)))))))))... ( -34.20) >DroSim_CAF1 44921 113 + 1 ACCACUAAUUGCGCAAAUUAUUUGGAGUUAUUUCCUAAUUUGUAUUCUCUGCUGACCGAGAUAGUUGGCAGCCAUUCAUUUCAGUCAGCAGCACCGAACCAGGCCAAUUAAUA .....(((((((((((((((...((((....)))))))))))).....((((((((.(((((...(((...)))...))))).))))))))...........).))))))... ( -30.60) >DroEre_CAF1 46852 112 + 1 ACCACUAAUUGCGCA-AUUAUUUGGAGUAAUUUCCUAAUUUGUAUUCUCUGCUGACCGAGAUAGUUGGCAGCCUUUCAUUUCAGUCAGCAGCACCGAACCAGGCCAAUUAAUA .....((((((.((.-.......((((....)))).............((((((((.(((((.(..((...))..).))))).))))))))...........))))))))... ( -27.40) >DroYak_CAF1 44077 112 + 1 ACCACUAAUUGCGCA-AUUAUUUGGAGUAAUUUCCUAAUUUGUAUUCUCUGCUGACCGAGAUAGUUGGCAGCCUUUCAUUUUAGUCAGCAGCACCGAACCAGGCCAAUUAAUA .....((((((.((.-.......((((....)))).............((((((((..((((.(..((...))..).))))..))))))))...........))))))))... ( -26.30) >consensus ACCACUAAUUGCGCAAAUUAUUUGGAGUAAUUUCCUAAUUUGUAUUCUCUGCUGACCGAGAUAGUUGGCAGCCAUUCAUUUCAGUCAGCAGCACCGAACCAGGCCAAUUAAUA .....((((((.((((((((...((((....)))))))))))).....((((((((.(((((...(((...)))...))))).))))))))..((......)).))))))... (-27.10 = -27.94 + 0.84)

| Location | 17,879,534 – 17,879,647 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 97.17 |
| Mean single sequence MFE | -33.70 |
| Consensus MFE | -31.86 |
| Energy contribution | -32.66 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.95 |
| SVM decision value | 4.27 |
| SVM RNA-class probability | 0.999856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17879534 113 - 20766785 UAUUAAUUGGCCUGGUUCGGUGGUGCUGACUGAAAUGAAUGGCUGCCAACUAUCUCGGUCAGCAGAGAAUACAAAUUAGGAAAUUACUCCAAAUAAUUUGCACAAUUAGUGGU ((((((((((((......)))..((((((((((.(((..(((...)))..))).)))))))))).......((((((((((......)))...)))))))..))))))))).. ( -32.20) >DroSec_CAF1 44443 113 - 1 UAUUAAUUGGCCUGGUUCGGCGCUGCUGACUGAAAUGAAUGGCUGCCAACUAUCUCGGUCAGCAGAGAAUACAAAUUAGGAAAUUACUCCAAAUAAUUUGCGCAAUUAGUGGU ((((((((((((......))).(((((((((((.(((..(((...)))..))).)))))))))))......((((((((((......)))...)))))))..))))))))).. ( -36.80) >DroSim_CAF1 44921 113 - 1 UAUUAAUUGGCCUGGUUCGGUGCUGCUGACUGAAAUGAAUGGCUGCCAACUAUCUCGGUCAGCAGAGAAUACAAAUUAGGAAAUAACUCCAAAUAAUUUGCGCAAUUAGUGGU (((((((((.....((((....(((((((((((.(((..(((...)))..))).))))))))))).)))).((((((((((......)))...)))))))..))))))))).. ( -35.70) >DroEre_CAF1 46852 112 - 1 UAUUAAUUGGCCUGGUUCGGUGCUGCUGACUGAAAUGAAAGGCUGCCAACUAUCUCGGUCAGCAGAGAAUACAAAUUAGGAAAUUACUCCAAAUAAU-UGCGCAAUUAGUGGU (((((((((((...((((....(((((((((((...((.((........)).))))))))))))).))))........(((......))).......-.)).))))))))).. ( -33.80) >DroYak_CAF1 44077 112 - 1 UAUUAAUUGGCCUGGUUCGGUGCUGCUGACUAAAAUGAAAGGCUGCCAACUAUCUCGGUCAGCAGAGAAUACAAAUUAGGAAAUUACUCCAAAUAAU-UGCGCAAUUAGUGGU (((((((((((...((((....(((((((((.....((.((........)).))..))))))))).))))........(((......))).......-.)).))))))))).. ( -30.00) >consensus UAUUAAUUGGCCUGGUUCGGUGCUGCUGACUGAAAUGAAUGGCUGCCAACUAUCUCGGUCAGCAGAGAAUACAAAUUAGGAAAUUACUCCAAAUAAUUUGCGCAAUUAGUGGU (((((((((((...((((....(((((((((((.(((..(((...)))..))).))))))))))).))))........(((......))).........)).))))))))).. (-31.86 = -32.66 + 0.80)

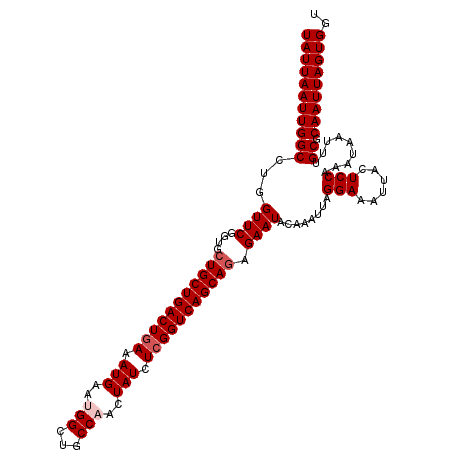

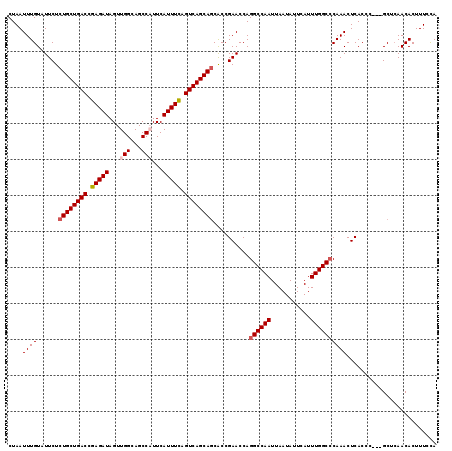
| Location | 17,879,568 – 17,879,684 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 96.08 |
| Mean single sequence MFE | -27.29 |
| Consensus MFE | -24.34 |
| Energy contribution | -24.98 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.31 |
| SVM RNA-class probability | 0.998977 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17879568 116 + 20766785 CUAAUUUGUAUUCUCUGCUGACCGAGAUAGUUGGCAGCCAUUCAUUUCAGUCAGCACCACCGAACCAGGCCAAUUAAUAUUCAUUUGGCCCAAACUCACCC---GCUGAACACUUUCCA .......((.(((..(((((((.(((((...(((...)))...))))).)))))))...........((((((...........))))))...........---...))).))...... ( -26.50) >DroSec_CAF1 44477 119 + 1 CUAAUUUGUAUUCUCUGCUGACCGAGAUAGUUGGCAGCCAUUCAUUUCAGUCAGCAGCGCCGAACCAGGCCAAUUAAUAUUCAUUUGGCCCAAACUCACCCGCCGCUCAACACUUUCCA ......(((......(((((((.(((((...(((...)))...))))).)))))))(((.((.....((((((...........))))))..........)).)))...)))....... ( -28.36) >DroSim_CAF1 44955 116 + 1 CUAAUUUGUAUUCUCUGCUGACCGAGAUAGUUGGCAGCCAUUCAUUUCAGUCAGCAGCACCGAACCAGGCCAAUUAAUAUUCAUUUGGCCCAAACUCACCC---GCUCAACACUUUCUA ....((((......((((((((.(((((...(((...)))...))))).))))))))..........((((((...........)))))))))).......---............... ( -27.90) >DroEre_CAF1 46885 116 + 1 CUAAUUUGUAUUCUCUGCUGACCGAGAUAGUUGGCAGCCUUUCAUUUCAGUCAGCAGCACCGAACCAGGCCAAUUAAUAUUCAUUUGGCCCAAACUCACCC---GCUCAACACUUUCCA ....((((......((((((((.(((((.(..((...))..).))))).))))))))..........((((((...........)))))))))).......---............... ( -26.70) >DroYak_CAF1 44110 115 + 1 CUAAUUUGUAUUCUCUGCUGACCGAGAUAGUUGGCAGCCUUUCAUUUUAGUCAGCAGCACCGAACCAGGCCAAUUAAUAUUCAUUUGGC-CAAACUCACCC---GCUCAACACUUUGCA ......((((....((((((((..((((.(..((...))..).))))..))))))))..........((((((...........)))))-)..........---...........)))) ( -27.00) >consensus CUAAUUUGUAUUCUCUGCUGACCGAGAUAGUUGGCAGCCAUUCAUUUCAGUCAGCAGCACCGAACCAGGCCAAUUAAUAUUCAUUUGGCCCAAACUCACCC___GCUCAACACUUUCCA ....((((......((((((((.(((((...(((...)))...))))).))))))))..........((((((...........))))))))))......................... (-24.34 = -24.98 + 0.64)


| Location | 17,879,568 – 17,879,684 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 96.08 |
| Mean single sequence MFE | -36.80 |
| Consensus MFE | -31.30 |
| Energy contribution | -32.14 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.29 |
| SVM RNA-class probability | 0.991802 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17879568 116 - 20766785 UGGAAAGUGUUCAGC---GGGUGAGUUUGGGCCAAAUGAAUAUUAAUUGGCCUGGUUCGGUGGUGCUGACUGAAAUGAAUGGCUGCCAACUAUCUCGGUCAGCAGAGAAUACAAAUUAG ......((((((.((---.(.((((.(..((((((.(((...))).))))))..))))).).))(((((((((.(((..(((...)))..))).)))))))))...))))))....... ( -39.00) >DroSec_CAF1 44477 119 - 1 UGGAAAGUGUUGAGCGGCGGGUGAGUUUGGGCCAAAUGAAUAUUAAUUGGCCUGGUUCGGCGCUGCUGACUGAAAUGAAUGGCUGCCAACUAUCUCGGUCAGCAGAGAAUACAAAUUAG ......(((((.((((.((((.....(..((((((.(((...))).))))))..))))).))))(((((((((.(((..(((...)))..))).)))))))))....)))))....... ( -41.60) >DroSim_CAF1 44955 116 - 1 UAGAAAGUGUUGAGC---GGGUGAGUUUGGGCCAAAUGAAUAUUAAUUGGCCUGGUUCGGUGCUGCUGACUGAAAUGAAUGGCUGCCAACUAUCUCGGUCAGCAGAGAAUACAAAUUAG ......(((((..((---.((.....(..((((((.(((...))).))))))..).)).)).(((((((((((.(((..(((...)))..))).)))))))))))..)))))....... ( -37.30) >DroEre_CAF1 46885 116 - 1 UGGAAAGUGUUGAGC---GGGUGAGUUUGGGCCAAAUGAAUAUUAAUUGGCCUGGUUCGGUGCUGCUGACUGAAAUGAAAGGCUGCCAACUAUCUCGGUCAGCAGAGAAUACAAAUUAG ......(((((..((---.((.....(..((((((.(((...))).))))))..).)).)).(((((((((((...((.((........)).)))))))))))))..)))))....... ( -35.70) >DroYak_CAF1 44110 115 - 1 UGCAAAGUGUUGAGC---GGGUGAGUUUG-GCCAAAUGAAUAUUAAUUGGCCUGGUUCGGUGCUGCUGACUAAAAUGAAAGGCUGCCAACUAUCUCGGUCAGCAGAGAAUACAAAUUAG ......(((((....---.(.(((((..(-(((((.(((...))).))))))..))))).).(((((((((.....((.((........)).))..)))))))))..)))))....... ( -30.40) >consensus UGGAAAGUGUUGAGC___GGGUGAGUUUGGGCCAAAUGAAUAUUAAUUGGCCUGGUUCGGUGCUGCUGACUGAAAUGAAUGGCUGCCAACUAUCUCGGUCAGCAGAGAAUACAAAUUAG ......(((((........(.((((...(((((((.(((...))).)))))))..)))).).(((((((((((.(((..(((...)))..))).)))))))))))..)))))....... (-31.30 = -32.14 + 0.84)

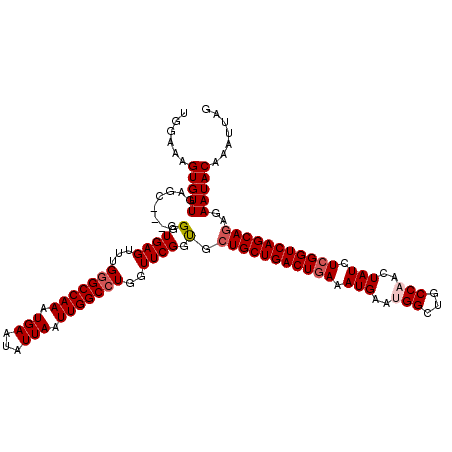
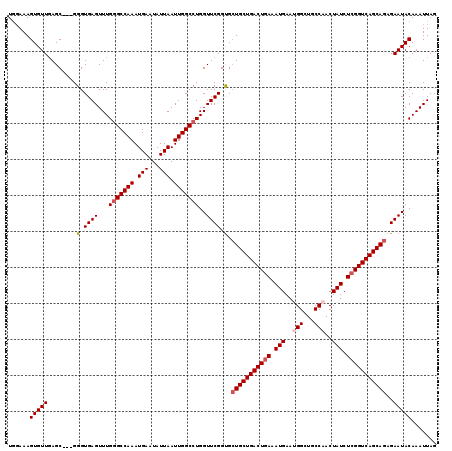
| Location | 17,879,607 – 17,879,718 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 95.19 |
| Mean single sequence MFE | -25.14 |
| Consensus MFE | -20.18 |
| Energy contribution | -20.42 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.874026 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17879607 111 + 20766785 AUUCAUUUCAGUCAGCACCACCGAACCAGGCCAAUUAAUAUUCAUUUGGCCCAAACUCACCC---GCUGAACACUUUCCAGCUGGGUGGAAAAUCCUUUAUUUAGCGAGCAACU ..........((..((............((((((...........)))))).....((((((---((((.........)))).))))))...............))..)).... ( -27.10) >DroSec_CAF1 44516 114 + 1 AUUCAUUUCAGUCAGCAGCGCCGAACCAGGCCAAUUAAUAUUCAUUUGGCCCAAACUCACCCGCCGCUCAACACUUUCCAGCUGGGUGGAAAAUCCUUUAUUUAGCGAGCAACU ..............((..(((..((...((((((...........)))))).....(((((((..(((...........))))))))))...........))..))).)).... ( -24.90) >DroSim_CAF1 44994 111 + 1 AUUCAUUUCAGUCAGCAGCACCGAACCAGGCCAAUUAAUAUUCAUUUGGCCCAAACUCACCC---GCUCAACACUUUCUAGCUGGGUGGAAAAUCCUUUAUUUAGCGAGCAACU ..............((.((.........((((((...........)))))).....((((((---(((...........))).))))))...............))..)).... ( -23.80) >DroEre_CAF1 46924 111 + 1 UUUCAUUUCAGUCAGCAGCACCGAACCAGGCCAAUUAAUAUUCAUUUGGCCCAAACUCACCC---GCUCAACACUUUCCAGCUGGGUGGAAAAUCCUUUAUUUAGCGAGCAACU ..............((.((.........((((((...........)))))).....((((((---(((...........))).))))))...............))..)).... ( -23.90) >DroYak_CAF1 44149 110 + 1 UUUCAUUUUAGUCAGCAGCACCGAACCAGGCCAAUUAAUAUUCAUUUGGC-CAAACUCACCC---GCUCAACACUUUGCAGCUGGCUGAAAAAUCCUUUAUUUAGCGAGCAACU .....(((((((((((............((((((...........)))))-)..........---((..........)).)))))))))))....................... ( -26.00) >consensus AUUCAUUUCAGUCAGCAGCACCGAACCAGGCCAAUUAAUAUUCAUUUGGCCCAAACUCACCC___GCUCAACACUUUCCAGCUGGGUGGAAAAUCCUUUAUUUAGCGAGCAACU ..........((..((............((((((...........)))))).....((((((...(((...........))).))))))...............))..)).... (-20.18 = -20.42 + 0.24)

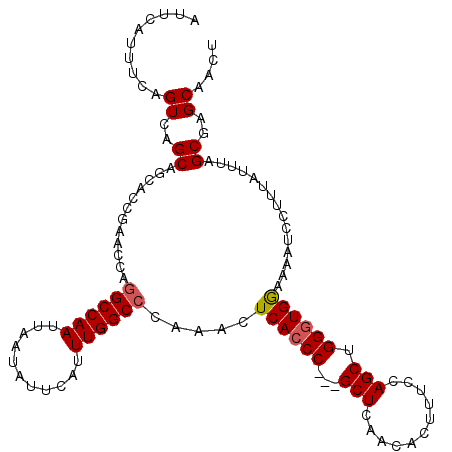

| Location | 17,879,607 – 17,879,718 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 95.19 |
| Mean single sequence MFE | -33.48 |
| Consensus MFE | -26.66 |
| Energy contribution | -27.10 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.645495 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17879607 111 - 20766785 AGUUGCUCGCUAAAUAAAGGAUUUUCCACCCAGCUGGAAAGUGUUCAGC---GGGUGAGUUUGGGCCAAAUGAAUAUUAAUUGGCCUGGUUCGGUGGUGCUGACUGAAAUGAAU ((((((((((..(((...........(((((.((((((.....))))))---))))).....(((((((.(((...))).))))))).)))..)))).)).))))......... ( -35.80) >DroSec_CAF1 44516 114 - 1 AGUUGCUCGCUAAAUAAAGGAUUUUCCACCCAGCUGGAAAGUGUUGAGCGGCGGGUGAGUUUGGGCCAAAUGAAUAUUAAUUGGCCUGGUUCGGCGCUGCUGACUGAAAUGAAU ..(..((((((......((.((((((((......)))))))).))....))))))..)..(..((((((.(((...))).))))))..).(((((...)))))........... ( -34.50) >DroSim_CAF1 44994 111 - 1 AGUUGCUCGCUAAAUAAAGGAUUUUCCACCCAGCUAGAAAGUGUUGAGC---GGGUGAGUUUGGGCCAAAUGAAUAUUAAUUGGCCUGGUUCGGUGCUGCUGACUGAAAUGAAU ..(..((((((.......((.....))...((((........)))))))---)))..)..(..((((((.(((...))).))))))..)((((((.(....)))))))...... ( -32.30) >DroEre_CAF1 46924 111 - 1 AGUUGCUCGCUAAAUAAAGGAUUUUCCACCCAGCUGGAAAGUGUUGAGC---GGGUGAGUUUGGGCCAAAUGAAUAUUAAUUGGCCUGGUUCGGUGCUGCUGACUGAAAUGAAA ..(..((((((......((.((((((((......)))))))).)).)))---)))..)..(..((((((.(((...))).))))))..)((((((.(....)))))))...... ( -35.10) >DroYak_CAF1 44149 110 - 1 AGUUGCUCGCUAAAUAAAGGAUUUUUCAGCCAGCUGCAAAGUGUUGAGC---GGGUGAGUUUG-GCCAAAUGAAUAUUAAUUGGCCUGGUUCGGUGCUGCUGACUAAAAUGAAA ..(..((((((......((.(((((.(((....))).))))).)).)))---)))..)....(-(((((.(((...))).))))))((((.(((.....)))))))........ ( -29.70) >consensus AGUUGCUCGCUAAAUAAAGGAUUUUCCACCCAGCUGGAAAGUGUUGAGC___GGGUGAGUUUGGGCCAAAUGAAUAUUAAUUGGCCUGGUUCGGUGCUGCUGACUGAAAUGAAU ....(((((((......((.((((((((......)))))))).))........)))))))..(((((((.(((...))).)))))))(((.((((...)))))))......... (-26.66 = -27.10 + 0.44)

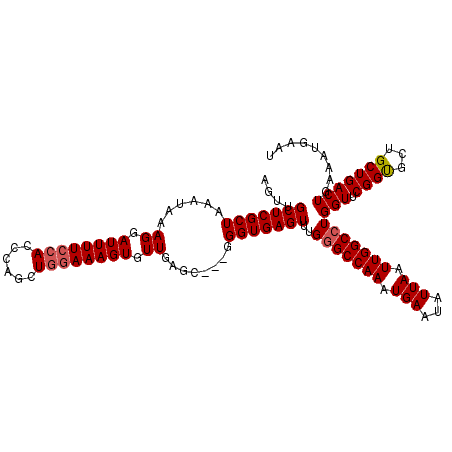
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:50:05 2006