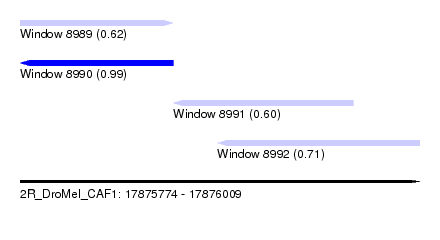
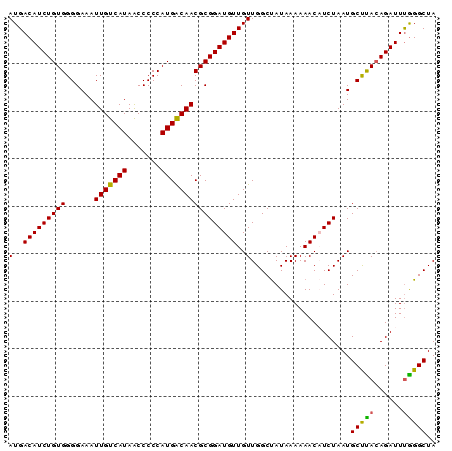
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,875,774 – 17,876,009 |
| Length | 235 |
| Max. P | 0.988571 |

| Location | 17,875,774 – 17,875,864 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -22.22 |
| Consensus MFE | -18.58 |
| Energy contribution | -18.10 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.619964 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17875774 90 + 20766785 UAGCCUGAAUCUGUAAGCAUUAGAUGUUUUUUAUAGCCAACAACAUCCGCGUUGUCAUGGGGGUUAUGACAAUUUCCCCACAGAUGUCAU .....(((((((((..((..((((.....))))..))..(((((......)))))....((((..((....))..)))))))))).))). ( -20.00) >DroSec_CAF1 40629 90 + 1 UAGCCCAAAUCUGUAAGCAUUAGAUGUUUUUUAUAGCCAACAACAUCCGCGUUGUCAUGGGGGUUAUGACAAUUUCCCCACAGAUGUCAU ((((((....((((((((((...))))...))))))((((((((......)))))..)))))))))(((((.(((......)))))))). ( -20.40) >DroSim_CAF1 41089 90 + 1 UAGCCCAAAUCUGUAAGCAUUAGAUGUUUUUUAUAGCCAACAACAUCCGCGUUGUCAUGGGGGUUAUGACAAUUUCCCCACAGAUGUCAU ((((((....((((((((((...))))...))))))((((((((......)))))..)))))))))(((((.(((......)))))))). ( -20.40) >DroEre_CAF1 43040 90 + 1 UAGCCCAAAUCUGUAGGCAUUAGAGGUUUUUUAUGGCCAACAACAUCCGCGUUGCCAUGGGGGCUAUGACAAUUUCCCCACAGAUGUCAU ((((((((((((.(((...))).)))))).(((((((.(((.........))))))))))))))))(((((.(((......)))))))). ( -24.90) >DroYak_CAF1 39776 90 + 1 UAGCCCAAAUCUUUGGGCAUUAGAGGUUUUUUAUAGCCAACAACAUCCGCGUUGUCAUGGGGGUUAUGACAAUUUCCCCACAGAUGUCAU ..((((((....))))))......((((......)))).(((((......)))))..((((((..((....))..)))).))........ ( -25.40) >consensus UAGCCCAAAUCUGUAAGCAUUAGAUGUUUUUUAUAGCCAACAACAUCCGCGUUGUCAUGGGGGUUAUGACAAUUUCCCCACAGAUGUCAU ((((((....(((((((.(........).)))))))((((((((......)))))..)))))))))(((((.(((......)))))))). (-18.58 = -18.10 + -0.48)

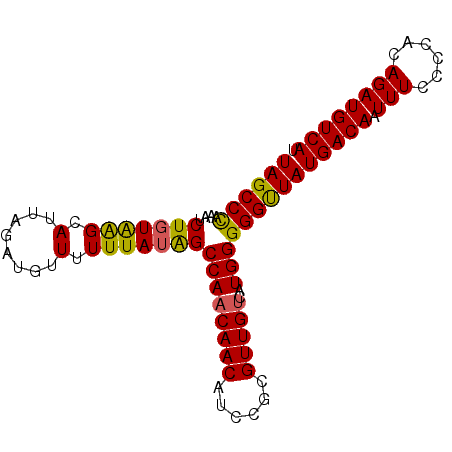
| Location | 17,875,774 – 17,875,864 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -23.88 |
| Consensus MFE | -22.38 |
| Energy contribution | -21.86 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988571 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17875774 90 - 20766785 AUGACAUCUGUGGGGAAAUUGUCAUAACCCCCAUGACAACGCGGAUGUUGUUGGCUAUAAAAAACAUCUAAUGCUUACAGAUUCAGGCUA .(((.(((((((((.(..(((((((.......)))))))...(((((((.((.......)).)))))))..).))))))))))))..... ( -24.20) >DroSec_CAF1 40629 90 - 1 AUGACAUCUGUGGGGAAAUUGUCAUAACCCCCAUGACAACGCGGAUGUUGUUGGCUAUAAAAAACAUCUAAUGCUUACAGAUUUGGGCUA .....(((((((((.(..(((((((.......)))))))...(((((((.((.......)).)))))))..).)))))))))........ ( -21.90) >DroSim_CAF1 41089 90 - 1 AUGACAUCUGUGGGGAAAUUGUCAUAACCCCCAUGACAACGCGGAUGUUGUUGGCUAUAAAAAACAUCUAAUGCUUACAGAUUUGGGCUA .....(((((((((.(..(((((((.......)))))))...(((((((.((.......)).)))))))..).)))))))))........ ( -21.90) >DroEre_CAF1 43040 90 - 1 AUGACAUCUGUGGGGAAAUUGUCAUAGCCCCCAUGGCAACGCGGAUGUUGUUGGCCAUAAAAAACCUCUAAUGCCUACAGAUUUGGGCUA (..((((((((((((.............))))..(....)))))))))..).....................(((((......))))).. ( -24.92) >DroYak_CAF1 39776 90 - 1 AUGACAUCUGUGGGGAAAUUGUCAUAACCCCCAUGACAACGCGGAUGUUGUUGGCUAUAAAAAACCUCUAAUGCCCAAAGAUUUGGGCUA (..(((((((((......(((((((.......))))))))))))))))..).....................((((((....)))))).. ( -26.50) >consensus AUGACAUCUGUGGGGAAAUUGUCAUAACCCCCAUGACAACGCGGAUGUUGUUGGCUAUAAAAAACAUCUAAUGCUUACAGAUUUGGGCUA (..(((((((((......(((((((.......))))))))))))))))..).....................(((((......))))).. (-22.38 = -21.86 + -0.52)

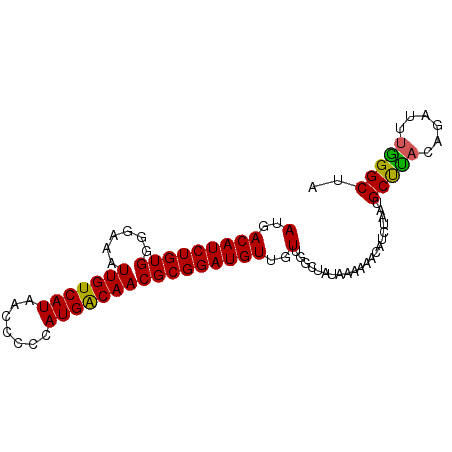
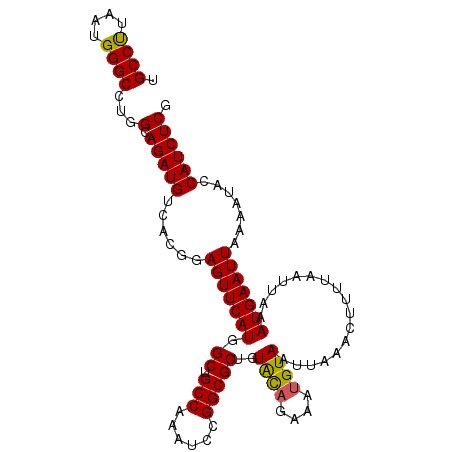
| Location | 17,875,864 – 17,875,970 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 95.28 |
| Mean single sequence MFE | -25.28 |
| Consensus MFE | -23.10 |
| Energy contribution | -22.74 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.604939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17875864 106 - 20766785 UGCCUUAAUGGGCCUGGCAGAUGUCACCGAGUUCAUGGCUGCCAAAUCCGGCGCUGUACAGAAAUGUAAUUAAACUUUUAAUUAAAUGAAUUAAAAUACCAUCUCG .((((....))))...(.(((((......((((((((((.(((......)))))).((((....)))).................))))))).......)))))). ( -25.22) >DroSec_CAF1 40719 106 - 1 UGCCUUAAUGGGCCUGGCAGAUGUCACGGAGUUCAUGGCUGCCAAAUCCGGCGCUGUACAGAAAUGUAAUUAAACUUUUAACUAAAUGAAUUAAAAUACCAUCUCG .((((....))))...(.(((((......((((((((((.(((......)))))).((((....)))).................))))))).......)))))). ( -25.22) >DroSim_CAF1 41179 106 - 1 UGCCCUAAUGGGCCUGGCAGAUGUCACGGAGUUCAUGGCUGCCAAAUCCGGCGCUGUACAGAAAUGUAAUUAAACUUUUAAUUAAAUGAAUUAAAAUACCAUCUCG .((((....))))...(.(((((......((((((((((.(((......)))))).((((....)))).................))))))).......)))))). ( -27.62) >DroEre_CAF1 43130 106 - 1 UGCCCUAAUGGGCCUGGCAGAUGUCACGGAGUUCAUGGCCGCCAAAUCUGGCGCAGUGUUGAAAUGUAAUUAAACUUUUAAUUAAAUGAAUUAAAAUACCAUCUCG .((((....))))...(.(((((......(((((((.((.((((....))))))...(((((((.((......)))))))))...))))))).......)))))). ( -25.92) >DroYak_CAF1 39866 106 - 1 UGCCUUAAUGGGCCUGGCAGAUGUCACGGAGUUCAUGGCCGCCAAAACCGGCGCAGUACACAAAUGUAAUUAAACUUUUAAUUAAAUGAAUUAAAAUACCAUCUCG .((((....))))...(.(((((......(((((((.((.(((......)))))............(((((((....))))))).))))))).......)))))). ( -22.42) >consensus UGCCUUAAUGGGCCUGGCAGAUGUCACGGAGUUCAUGGCUGCCAAAUCCGGCGCUGUACAGAAAUGUAAUUAAACUUUUAAUUAAAUGAAUUAAAAUACCAUCUCG .((((....))))...(.(((((......(((((((.((.(((......)))))..((((....)))).................))))))).......)))))). (-23.10 = -22.74 + -0.36)


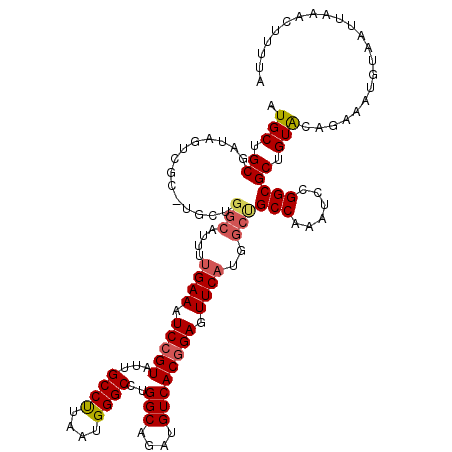
| Location | 17,875,890 – 17,876,009 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.02 |
| Mean single sequence MFE | -36.18 |
| Consensus MFE | -26.46 |
| Energy contribution | -26.94 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713270 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17875890 119 - 20766785 AUGCUGCGAUAUUCGC-UGCUGGCAUUUUGAAAUCCGUAUUGCCUUAAUGGGCCUGGCAGAUGUCACCGAGUUCAUGGCUGCCAAAUCCGGCGCUGUACAGAAAUGUAAUUAAACUUUUA ..((.(((.....)))-.))..(((((((((.(((.((...((((....))))...)).))).)).........(((((.(((......))))))))...)))))))............. ( -30.70) >DroSec_CAF1 40745 119 - 1 AUGCUGCGAUAGUCGC-UGCUGGCAUUUUGAAAUCCGUAUUGCCUUAAUGGGCCUGGCAGAUGUCACGGAGUUCAUGGCUGCCAAAUCCGGCGCUGUACAGAAAUGUAAUUAAACUUUUA ...(((..((((.(((-((.((((((..((((.(((((...((((....))))..(((....)))))))).))))..).)))))....))))))))).)))................... ( -36.90) >DroSim_CAF1 41205 119 - 1 AUGCUGCGAUAGUCGC-UGCUGGCAUUUUGAAAUCCGUAUUGCCCUAAUGGGCCUGGCAGAUGUCACGGAGUUCAUGGCUGCCAAAUCCGGCGCUGUACAGAAAUGUAAUUAAACUUUUA ...(((..((((.(((-((.((((((..((((.(((((...((((....))))..(((....)))))))).))))..).)))))....))))))))).)))................... ( -39.30) >DroEre_CAF1 43156 98 - 1 AUGCUGC----------------------GAAAUCCGUAUUGCCCUAAUGGGCCUGGCAGAUGUCACGGAGUUCAUGGCCGCCAAAUCUGGCGCAGUGUUGAAAUGUAAUUAAACUUUUA (((((((----------------------(((.(((((...((((....))))..(((....)))))))).)))......((((....)))))))))))..................... ( -32.80) >DroYak_CAF1 39892 120 - 1 AUGCUGCGUUAGUCGUUUGCUGGCAUUUUGAAAUCCGUAUUGCCUUAAUGGGCCUGGCAGAUGUCACGGAGUUCAUGGCCGCCAAAACCGGCGCAGUACACAAAUGUAAUUAAACUUUUA .((((((((..((..((((.((((....((((.(((((...((((....))))..(((....)))))))).))))..)))).))))))..))))))))...................... ( -41.20) >consensus AUGCUGCGAUAGUCGC_UGCUGGCAUUUUGAAAUCCGUAUUGCCUUAAUGGGCCUGGCAGAUGUCACGGAGUUCAUGGCUGCCAAAUCCGGCGCUGUACAGAAAUGUAAUUAAACUUUUA .(((.((..............(((....((((.(((((...((((....))))..(((....)))))))).))))..)))(((......))))).)))...................... (-26.46 = -26.94 + 0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:57 2006