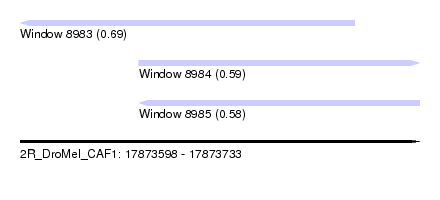
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,873,598 – 17,873,733 |
| Length | 135 |
| Max. P | 0.686634 |

| Location | 17,873,598 – 17,873,711 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 94.34 |
| Mean single sequence MFE | -25.61 |
| Consensus MFE | -22.20 |
| Energy contribution | -22.72 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.686634 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17873598 113 - 20766785 GUUAUUGGCUUUUUAUCUGGUCCAAACGCUUUAAAUGCAGAUUUAUGCAAUUCCCGUUUGAUGGGGAAUAUUAUUUGUGGACCGAUCCCAAAGACCAUUGAAUUCAAUCGAAA .(((.(((((((..(((.((((((...........((((......))))((((((........))))))........)))))))))...)))).))).)))............ ( -29.50) >DroSec_CAF1 38516 113 - 1 GUUAUUGGCUUUUUAUCUGGUCCAAACGGUUUAAAUGCAGAUUUAUGCAAUUCCCGUUUGAUGGGGAAUAUUAUUUGUGGACCGAUCCCAAAGACCAUUGAAUUCAAUCGAAA .(((.(((((((..(((.((((((...........((((......))))((((((........))))))........)))))))))...)))).))).)))............ ( -29.50) >DroSim_CAF1 38955 113 - 1 GUUAUUGGCUUUUUAUCUGGUCCAAACGCUUUAAAUGCAGAUUUAUGCAAUUCCCGUUUGAUGGGGAAUAUUAUUUGUGGAUCGAUCCCAAAGACCAUUGAAUUCAAUCGAAA .(((.(((((((..(((.((((((...........((((......))))((((((........))))))........)))))))))...)))).))).)))............ ( -26.90) >DroEre_CAF1 40910 113 - 1 GUUAUUGGCAUUUUAUCUGGCCCAAACGCUUUAAAUGCAGAUUUAUGCAAUUCCCAUUUGAUGGCGAAUAAUAUUUGUGGCCCCCUUCCAAAGACCAUUGAAUUCAAUCGAAA (((.((((..........((((((((.........((((......))))((((((((...)))).))))....)))).)))).....)))).)))..((((......)))).. ( -21.76) >DroYak_CAF1 37649 113 - 1 GUUAUUGGCUUUUUAUCUGGCCCAAACGCUUUAAAUGCAGAUUUAUGCAAUUCCCAUUUGAUGGCAAAUAUUAUUUGUGGUCCAAUCCCAAAGACCAUUGAAUUGAAUCGAAA (((...((((........))))..)))((.......)).((((((..(((....((..(((((.....)))))..))(((((..........))))))))...)))))).... ( -20.40) >consensus GUUAUUGGCUUUUUAUCUGGUCCAAACGCUUUAAAUGCAGAUUUAUGCAAUUCCCGUUUGAUGGGGAAUAUUAUUUGUGGACCGAUCCCAAAGACCAUUGAAUUCAAUCGAAA .(((.(((((((.....(((((((...........((((......)))).(((((((...)))))))..........))))))).....)))).))).)))............ (-22.20 = -22.72 + 0.52)

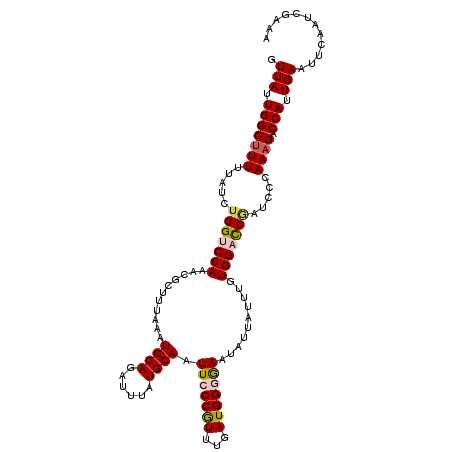
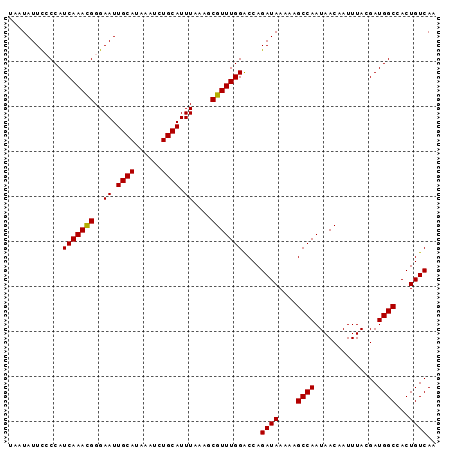
| Location | 17,873,638 – 17,873,733 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 96.42 |
| Mean single sequence MFE | -17.84 |
| Consensus MFE | -15.26 |
| Energy contribution | -15.02 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.588480 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17873638 95 + 20766785 UAAUAUUCCCCAUCAAACGGGAAUUGCAUAAAUCUGCAUUUAAAGCGUUUGGACCAGAUAAAAAGCCAAUAACAAUUUACGAUGGCCACUGUCAA ............(((((((..((.((((......)))).))....)))))))....((((....((((..............))))...)))).. ( -15.74) >DroSec_CAF1 38556 95 + 1 UAAUAUUCCCCAUCAAACGGGAAUUGCAUAAAUCUGCAUUUAAACCGUUUGGACCAGAUAAAAAGCCAAUAACAAUUUACGAUGGCCACUGUCAA ............((((((((.((.((((......)))).))...))))))))....((((....((((..............))))...)))).. ( -20.44) >DroSim_CAF1 38995 95 + 1 UAAUAUUCCCCAUCAAACGGGAAUUGCAUAAAUCUGCAUUUAAAGCGUUUGGACCAGAUAAAAAGCCAAUAACAAUUUACGAUGGCCACUGUCAA ............(((((((..((.((((......)))).))....)))))))....((((....((((..............))))...)))).. ( -15.74) >DroEre_CAF1 40950 95 + 1 UAUUAUUCGCCAUCAAAUGGGAAUUGCAUAAAUCUGCAUUUAAAGCGUUUGGGCCAGAUAAAAUGCCAAUAACAAUUUACGAUGGCCACUGUCAA ........((((((..........((((......))))..((((..(((..(((..........)))...)))..)))).))))))......... ( -18.70) >DroYak_CAF1 37689 95 + 1 UAAUAUUUGCCAUCAAAUGGGAAUUGCAUAAAUCUGCAUUUAAAGCGUUUGGGCCAGAUAAAAAGCCAAUAACAAUUUACGAUGGCCACUGUCAA ........((((((..........((((......))))..((((..(((..(((..........)))...)))..)))).))))))......... ( -18.60) >consensus UAAUAUUCCCCAUCAAACGGGAAUUGCAUAAAUCUGCAUUUAAAGCGUUUGGACCAGAUAAAAAGCCAAUAACAAUUUACGAUGGCCACUGUCAA ............(((((((..((.((((......)))).))....)))))))....((((....((((..............))))...)))).. (-15.26 = -15.02 + -0.24)

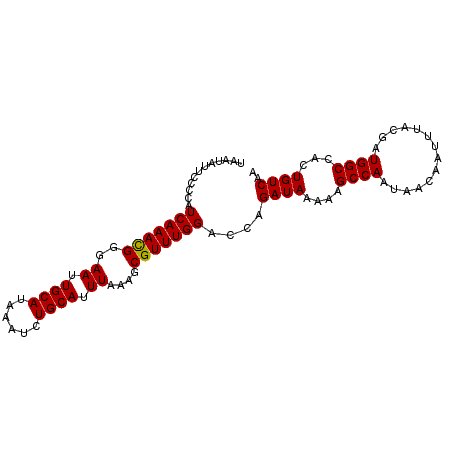
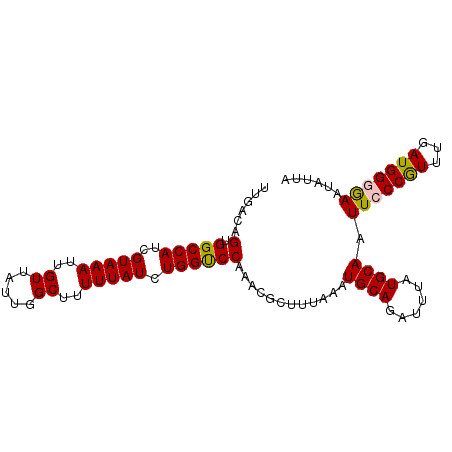
| Location | 17,873,638 – 17,873,733 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 96.42 |
| Mean single sequence MFE | -21.82 |
| Consensus MFE | -18.90 |
| Energy contribution | -18.66 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.576479 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17873638 95 - 20766785 UUGACAGUGGCCAUCGUAAAUUGUUAUUGGCUUUUUAUCUGGUCCAAACGCUUUAAAUGCAGAUUUAUGCAAUUCCCGUUUGAUGGGGAAUAUUA .......((((((..(((((..((.....))..))))).))).)))...........((((......))))((((((........)))))).... ( -20.80) >DroSec_CAF1 38556 95 - 1 UUGACAGUGGCCAUCGUAAAUUGUUAUUGGCUUUUUAUCUGGUCCAAACGGUUUAAAUGCAGAUUUAUGCAAUUCCCGUUUGAUGGGGAAUAUUA ..(((..((((((..(((((..((.....))..))))).))).)))....)))....((((......))))((((((........)))))).... ( -22.10) >DroSim_CAF1 38995 95 - 1 UUGACAGUGGCCAUCGUAAAUUGUUAUUGGCUUUUUAUCUGGUCCAAACGCUUUAAAUGCAGAUUUAUGCAAUUCCCGUUUGAUGGGGAAUAUUA .......((((((..(((((..((.....))..))))).))).)))...........((((......))))((((((........)))))).... ( -20.80) >DroEre_CAF1 40950 95 - 1 UUGACAGUGGCCAUCGUAAAUUGUUAUUGGCAUUUUAUCUGGCCCAAACGCUUUAAAUGCAGAUUUAUGCAAUUCCCAUUUGAUGGCGAAUAAUA ......(.(((((..(((((.(((.....))).))))).))))))...(((((((((((..((((.....))))..))))))).))))....... ( -23.00) >DroYak_CAF1 37689 95 - 1 UUGACAGUGGCCAUCGUAAAUUGUUAUUGGCUUUUUAUCUGGCCCAAACGCUUUAAAUGCAGAUUUAUGCAAUUCCCAUUUGAUGGCAAAUAUUA .........(((((((((((.((((...((((........))))..)))).))))..((((......)))).........)))))))........ ( -22.40) >consensus UUGACAGUGGCCAUCGUAAAUUGUUAUUGGCUUUUUAUCUGGUCCAAACGCUUUAAAUGCAGAUUUAUGCAAUUCCCGUUUGAUGGGGAAUAUUA ......(.(((((..(((((..((.....))..))))).))))))............((((......)))).(((((((...)))))))...... (-18.90 = -18.66 + -0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:51 2006