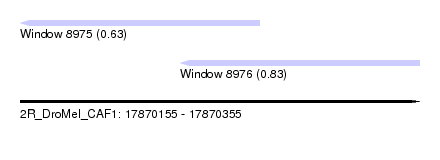
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,870,155 – 17,870,355 |
| Length | 200 |
| Max. P | 0.832723 |

| Location | 17,870,155 – 17,870,275 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.42 |
| Mean single sequence MFE | -27.61 |
| Consensus MFE | -20.64 |
| Energy contribution | -21.28 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.628925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17870155 120 - 20766785 UUACAAAUGGAGUGGGGAAAACAUUCUUACUUCAAGAAAAACUACUCAUUUUCAGCUGCUCUUAAGUACCUCUGCUCAUGCAUUUUGAAUGGCGAUGACGGAAAAACUUUGUCAUUUAAU .......(((((((((((.....))))))))))).(((((........))))).(((..((..((.......(((....)))))..))..)))(((((((((.....))))))))).... ( -25.31) >DroSec_CAF1 34989 120 - 1 UUACAAAUGCAGCGGGGAAAACAUUCUUAGUUUAAGAAAAACUACUCAUUUUCAGCUGCUCUUAAGUACCGCUGCUCAUGCAUUUUGAAUGGCGAUGACGGAAAAACUUUGUCAUUUAAU ........(((((((.(((((......((((((.....))))))....))))).(((.......))).))))))).........(((((((((((.............))))))))))). ( -28.52) >DroSim_CAF1 35346 120 - 1 UUUCAAAUGCAGCGGGAAAAACAUUCUUAGUUUAAGAAAAACUACUCAUUUUCAGCUGCUCUUAAGUACCGCUGCUCAUGCAUUUUGAAUGGCGAUGACGGAAAAACUUUGUCAUUUAAU .((((((.((((((((((((.......((((((.....))))))....))))).(((.......))).)))))))........))))))....(((((((((.....))))))))).... ( -28.00) >DroEre_CAF1 37455 110 - 1 ----------AGUGGGAAAAGCAUUCUUAGUUGAAGAAAAACUACUAAUUUUCAGCUGCUUUUAAGUACCGCUGCUCAUGCAUUUUGAAUGGCGAUGACGGGAAAAGUUUGUCAUUUAAU ----------(((((.(((((((......((((((((...........))))))))))))))).....)))))((....))...(((((((((((..((.......))))))))))))). ( -27.00) >DroYak_CAF1 33936 120 - 1 UUGCAAAUGCAGUGGCGAAAACAUUCUUAGUUAAAGAAAAACUACUCAUUUUCAGCUACAUUUAAGUACCGCUGCUCAUGCAUUUUGAAUGGCGAUGAGGGAAAAACUUUGUCAUUUAAU .((((...(((((((.(((((......(((((.......)))))....))))).(((.......))).)))))))...))))..(((((((((((...((......))))))))))))). ( -29.20) >consensus UUACAAAUGCAGUGGGGAAAACAUUCUUAGUUUAAGAAAAACUACUCAUUUUCAGCUGCUCUUAAGUACCGCUGCUCAUGCAUUUUGAAUGGCGAUGACGGAAAAACUUUGUCAUUUAAU ........(((((((........(((((.....)))))................(((.......))).))))))).........(((((((((((.............))))))))))). (-20.64 = -21.28 + 0.64)

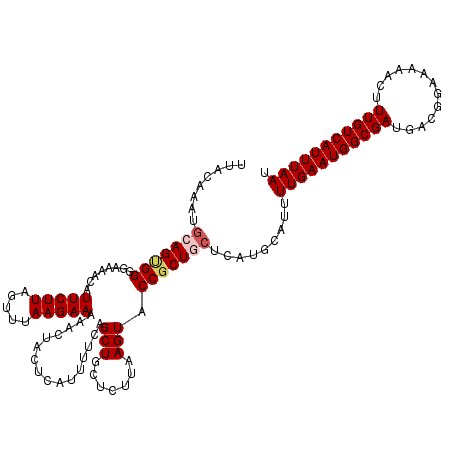
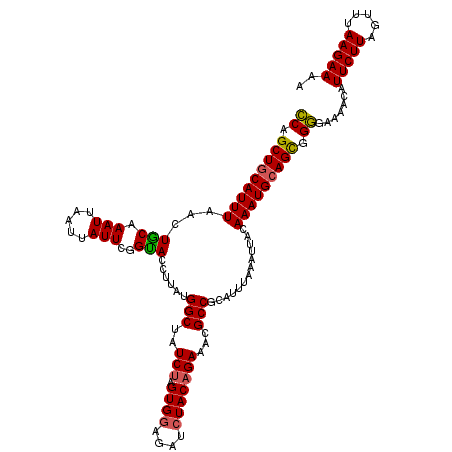
| Location | 17,870,235 – 17,870,355 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.06 |
| Mean single sequence MFE | -27.25 |
| Consensus MFE | -23.27 |
| Energy contribution | -23.02 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.832723 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17870235 120 - 20766785 CCAGCUGCAUUUAACUACAAAUUAAUUAUUCGGUACCUUAUGGCUAUCUAGUGGAGAUCUACAGAAACGCCGCAUUUAAAUUACAAAUGGAGUGGGGAAAACAUUCUUACUUCAAGAAAA ...((.((.......................((((((....)).))))..((((....))))......)).))..............(((((((((((.....)))))))))))...... ( -24.10) >DroSec_CAF1 35069 120 - 1 CCAGCUGCAUUUAACUGCAAAUUAAUUAUUCGGCACCUUAUGGCUAUCUAGUGGAGAUCUACAGAAACGCCGCAUUUAAAUUACAAAUGCAGCGGGGAAAACAUUCUUAGUUUAAGAAAA ((.((((((((....((.((.((((.....((((.((....))...(((.((((....)))))))...))))...)))).)).)))))))))).)).......((((((...)))))).. ( -29.00) >DroSim_CAF1 35426 120 - 1 CCAGCUGCAUUUAACUGCAAAUUAUUUAUUCGGUACCUUAUGGCUAUCUAGUGGAGAUCUACAGAAACGCCGCAUUUAAAUUUCAAAUGCAGCGGGAAAAACAUUCUUAGUUUAAGAAAA ((.(((((((((.((((.((........)))))).......(((..(((.((((....)))))))...))).............))))))))).)).......((((((...)))))).. ( -28.30) >DroYak_CAF1 34016 120 - 1 GCAGCUGCAUUUAACUGCAAAUUAAUUAUUGGGUACCUUAUGGCUAUCAAGUGAAGAUCUACAGAAAUGCCGCAUUUAAAUUGCAAAUGCAGUGGCGAAAACAUUCUUAGUUAAAGAAAA ((.((((((((....(((((.(((((((((.((((((....)).)))).))))).......((....))......)))).))))))))))))).)).......(((((.....))))).. ( -27.60) >consensus CCAGCUGCAUUUAACUGCAAAUUAAUUAUUCGGUACCUUAUGGCUAUCUAGUGGAGAUCUACAGAAACGCCGCAUUUAAAUUACAAAUGCAGCGGGGAAAACAUUCUUAGUUUAAGAAAA ((.(((((((((...(((.(((.....)))..)))......(((..(((.((((....)))))))...))).............))))))))).)).......(((((.....))))).. (-23.27 = -23.02 + -0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:42 2006