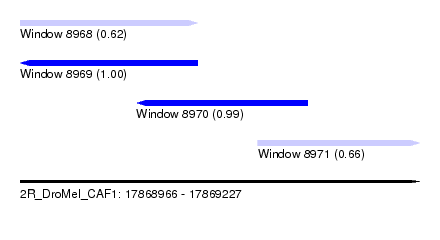
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,868,966 – 17,869,227 |
| Length | 261 |
| Max. P | 0.999068 |

| Location | 17,868,966 – 17,869,082 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.58 |
| Mean single sequence MFE | -22.58 |
| Consensus MFE | -17.38 |
| Energy contribution | -18.18 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.619334 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17868966 116 + 20766785 AGAAAAUAACUUAU----UUGCAUAGUAAAACGGGCAAUUGAAUGCCUUGAGAAUCCAAACAAUCGACAAACAAAGAACCCGUCUUGCUAUUCAUUUAUGGAAGUAUCCAUAUUUUCAGC .(((((((...((.----.((.(((((((.(((((...((((.((..(((......))).)).))))(.......)..))))).))))))).))..))((((....)))))))))))... ( -24.60) >DroSec_CAF1 33832 116 + 1 AGAAAAUAUCUUAU----UUGCAUAGUAAAACGGGCAAUUGAAUGCCUUGAGAACCCAAACAAUCGACAAACAAAGAACCCGUUUUGCUAUUCAUUUAUGGAAGUAUCCACAUUUCCAGC ...........((.----.((.(((((((((((((...((((.((..(((......))).)).))))(.......)..))))))))))))).))..))(((((((......))))))).. ( -26.00) >DroSim_CAF1 34183 116 + 1 AGAAAAUAUCUUAU----UUGCAUAGUAAAACGGGCAAUUGAAUGCCUUGAGAACCCAAACAAUCGACAAACAAAGAACCCGUCUUGCUAUUCAUUUAUGGAAGUAUCCACAUUUCCAGC ...........((.----.((.(((((((.(((((...((((.((..(((......))).)).))))(.......)..))))).))))))).))..))(((((((......))))))).. ( -22.50) >DroEre_CAF1 36259 120 + 1 AGAAAACAUCUUACAUAUUUACAUAAUAAAACGGGCAAUUGAAUGCCUUGAGAGCCCAAACAAUCGACAAACAAAGAACCCGUUUUGCUAUUCAUUUAUGGAAGUAUCCACAUUUUCAGC .(((((................(((.(((((((((...(((..((..((((..(......)..))))))..)))....))))))))).))).......((((....))))..)))))... ( -17.30) >DroYak_CAF1 32687 116 + 1 UAGAAAUAUCUUGC----UUACUUAAUAAAAGGGGCAAUUGAAUGCCUUGAGAGGCCAAACAAUCGACAAACAAAGAACCUGUUUUGCUAUUCAUUUAUGGAAGUAUCCACAUUUUCAGC ..((((.....((.----.(((((.(((((.((((((......))))))(((.(((.(((((.((..........))...))))).))).))).)))))..)))))..))...))))... ( -22.50) >consensus AGAAAAUAUCUUAU____UUGCAUAGUAAAACGGGCAAUUGAAUGCCUUGAGAACCCAAACAAUCGACAAACAAAGAACCCGUUUUGCUAUUCAUUUAUGGAAGUAUCCACAUUUUCAGC ......................(((((((((((((...((((.((..(((......))).)).))))(.......)..))))))))))))).......(((((((......))))))).. (-17.38 = -18.18 + 0.80)



| Location | 17,868,966 – 17,869,082 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.58 |
| Mean single sequence MFE | -29.74 |
| Consensus MFE | -28.16 |
| Energy contribution | -27.60 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.95 |
| SVM decision value | 3.35 |
| SVM RNA-class probability | 0.999068 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17868966 116 - 20766785 GCUGAAAAUAUGGAUACUUCCAUAAAUGAAUAGCAAGACGGGUUCUUUGUUUGUCGAUUGUUUGGAUUCUCAAGGCAUUCAAUUGCCCGUUUUACUAUGCAA----AUAAGUUAUUUUCU ...(((((((((((....))))))..((.((((.(((((((((...(((..((((....(........)....))))..)))..))))))))).)))).)).----........))))). ( -32.10) >DroSec_CAF1 33832 116 - 1 GCUGGAAAUGUGGAUACUUCCAUAAAUGAAUAGCAAAACGGGUUCUUUGUUUGUCGAUUGUUUGGGUUCUCAAGGCAUUCAAUUGCCCGUUUUACUAUGCAA----AUAAGAUAUUUUCU ...(((((((((((....))).....((.((((.(((((((((...(((..((((....(........)....))))..)))..))))))))).)))).)).----.....)))))))). ( -31.80) >DroSim_CAF1 34183 116 - 1 GCUGGAAAUGUGGAUACUUCCAUAAAUGAAUAGCAAGACGGGUUCUUUGUUUGUCGAUUGUUUGGGUUCUCAAGGCAUUCAAUUGCCCGUUUUACUAUGCAA----AUAAGAUAUUUUCU ...(((((((((((....))).....((.((((.(((((((((...(((..((((....(........)....))))..)))..))))))))).)))).)).----.....)))))))). ( -31.90) >DroEre_CAF1 36259 120 - 1 GCUGAAAAUGUGGAUACUUCCAUAAAUGAAUAGCAAAACGGGUUCUUUGUUUGUCGAUUGUUUGGGCUCUCAAGGCAUUCAAUUGCCCGUUUUAUUAUGUAAAUAUGUAAGAUGUUUUCU ...(((((((((((....)))))......((((.(((((((((...(((..((((((..((....))..))..))))..)))..))))))))).))))...............)))))). ( -29.80) >DroYak_CAF1 32687 116 - 1 GCUGAAAAUGUGGAUACUUCCAUAAAUGAAUAGCAAAACAGGUUCUUUGUUUGUCGAUUGUUUGGCCUCUCAAGGCAUUCAAUUGCCCCUUUUAUUAAGUAA----GCAAGAUAUUUCUA (((.....((((((....))))))((((((..((.((((((.....))))))...(((((....((((....))))...)))))))....)))))).....)----)).(((....))). ( -23.10) >consensus GCUGAAAAUGUGGAUACUUCCAUAAAUGAAUAGCAAAACGGGUUCUUUGUUUGUCGAUUGUUUGGGUUCUCAAGGCAUUCAAUUGCCCGUUUUACUAUGCAA____AUAAGAUAUUUUCU ...(((((((((((....)))))...((.((((.(((((((((...(((..((((....(........)....))))..)))..))))))))).)))).))............)))))). (-28.16 = -27.60 + -0.56)

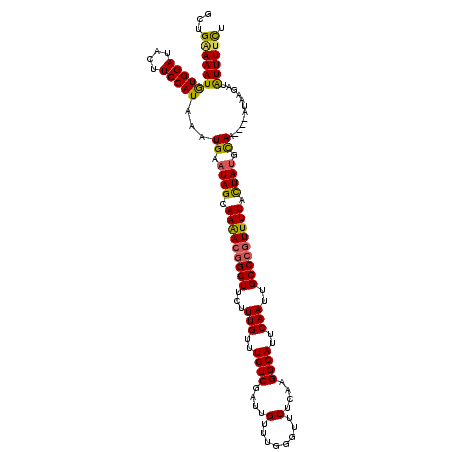
| Location | 17,869,042 – 17,869,154 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.41 |
| Mean single sequence MFE | -27.75 |
| Consensus MFE | -25.47 |
| Energy contribution | -24.95 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.991563 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17869042 112 - 20766785 AGAGCUUUCAGCUGGAAUGUUUGCUGGCAAGUG--------UACGUAAAUUAAAACAAGAGCCGCAAUCAAGUGUUCAUGGCUGAAAAUAUGGAUACUUCCAUAAAUGAAUAGCAAGACG ...((((((((((((((..((((.((((...((--------(............)))...))))....))))..))).))))))))).((((((....))))))........))...... ( -30.30) >DroSec_CAF1 33908 112 - 1 AGAGCUUUCAGCCGGAAUGUUUUCUCGCAAGUG--------UACGUAAAUUAAAACAAGAGCUGCAAUCAAGUGUUCAUGGCUGGAAAUGUGGAUACUUCCAUAAAUGAAUAGCAAAACG ...((((((((((((((..(((....(((.((.--------...((........))....)))))....)))..))).))))))))).((((((....))))))........))...... ( -28.50) >DroSim_CAF1 34259 112 - 1 AGAGCUUUCAGCUGGAAUGUUUGCUCGCAAGUG--------UACGUAAAUUAAAACAAGAGCCGCAAUCAAGUGUUCAUGGCUGGAAAUGUGGAUACUUCCAUAAAUGAAUAGCAAGACG ...((((((((((((((..((((...((..((.--------...((........))....)).))...))))..))).))))))))).((((((....))))))........))...... ( -26.00) >DroEre_CAF1 36339 112 - 1 AGAGCUUUCAGCUGGAAUGUUUGUUCGAAAGUG--------UACGCAAAUUAAAACAAGAGCCGCAAUCAAGUGUUCAUGGCUGAAAAUGUGGAUACUUCCAUAAAUGAAUAGCAAAACG ...((((((((((((((..((((..(....)((--------(..((..............)).)))..))))..))).))))))))).((((((....))))))........))...... ( -26.24) >DroYak_CAF1 32763 120 - 1 AGAGCUUGCAGCCGAAAUGUUUGUUCGGAAGUACACAUACAUACGUAAAUUAAAACAAGAGCCGCAAUCAAGUGUUCAUGGCUGAAAAUGUGGAUACUUCCAUAAAUGAAUAGCAAAACA ...(((..((((((..((((.(((.(....)...))).))))................((((.((......)))))).))))))....((((((....)))))).......)))...... ( -27.70) >consensus AGAGCUUUCAGCUGGAAUGUUUGCUCGCAAGUG________UACGUAAAUUAAAACAAGAGCCGCAAUCAAGUGUUCAUGGCUGAAAAUGUGGAUACUUCCAUAAAUGAAUAGCAAAACG ...(((((((((((...(((((........(((........)))........))))).((((.((......)))))).))))))))).((((((....))))))........))...... (-25.47 = -24.95 + -0.52)

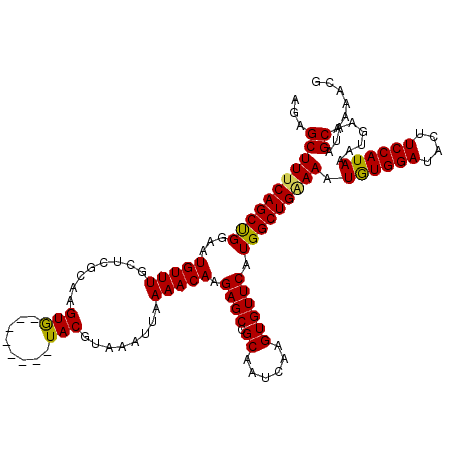
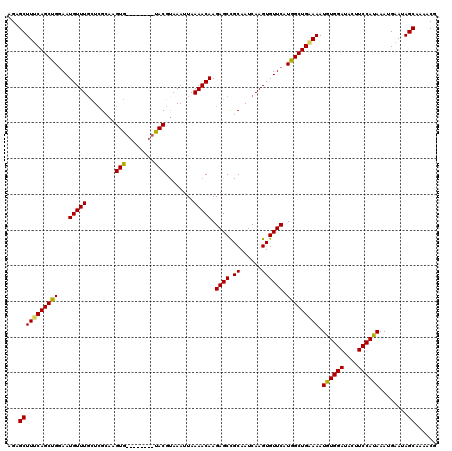
| Location | 17,869,121 – 17,869,227 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 89.43 |
| Mean single sequence MFE | -21.56 |
| Consensus MFE | -16.62 |
| Energy contribution | -16.94 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.655625 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17869121 106 + 20766785 -------CACUUGCCAGCAAACAUUCCAGCUGAAAGCUCUUUAAGCUGGAAAAACAAACAAAAAUCAGGCGGAUAAACGGACUUUAAAUUUUUUGAAUACGCAGCAAGGCAAA -------..(((((..((.....((((((((.(((....))).)))))))).......((((((..((.((......))..))......)))))).....)).)))))..... ( -24.60) >DroSec_CAF1 33987 106 + 1 -------CACUUGCGAGAAAACAUUCCGGCUGAAAGCUCUUUAAGCUGGAAAAACAAACAACAAUCAGACGGAUAAACGGACUUUAAAUUUUUUGAAUACGCAGCAUGGCAAA -------...(((((........((((((((.(((....))).))))))))...((((........((.((......))..))........))))....)))))......... ( -19.09) >DroSim_CAF1 34338 106 + 1 -------CACUUGCGAGCAAACAUUCCAGCUGAAAGCUCUUUAAGCUGGAAAAACAAACAAAAAUCAGACGGAUAAACGGACUUUAAAUUUUUUGAAUACGCAGCAUGGCAAA -------...((((.(((.....((((((((.(((....))).)))))))).......((((((..((.((......))..))......))))))........)).).)))). ( -22.20) >DroEre_CAF1 36418 106 + 1 -------CACUUUCGAACAAACAUUCCAGCUGAAAGCUCUUUAAGCUGGGAAGACAAACAAAAAUCAGACGGUAAAACGGACUUUAAAUUUUUUGAAUACGCAGAAAGGCAAA -------..(((((.........((((((((.(((....))).)))))))).......((((((......(((.......)))......))))))........)))))..... ( -20.40) >DroYak_CAF1 32843 113 + 1 GUAUGUGUACUUCCGAACAAACAUUUCGGCUGCAAGCUCUUUAAGCUGGAAAAACAAACAAAAAUCAGAGGAAUAAACGGACCUUAAAUUUUUUGAAUACGCAGCAGGGCAAA .....((..((..((((.......))))(((((.((((.....))))...........((((((...((((..........))))....)))))).....)))))))..)).. ( -21.50) >consensus _______CACUUGCGAGCAAACAUUCCAGCUGAAAGCUCUUUAAGCUGGAAAAACAAACAAAAAUCAGACGGAUAAACGGACUUUAAAUUUUUUGAAUACGCAGCAAGGCAAA .........(((((.........((((((((.(((....))).)))))))).......((((((.....((......))..........))))))........)))))..... (-16.62 = -16.94 + 0.32)

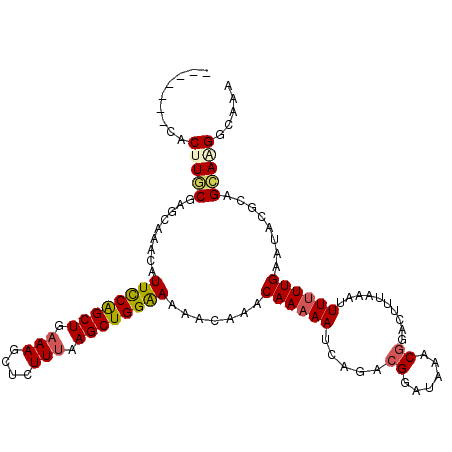
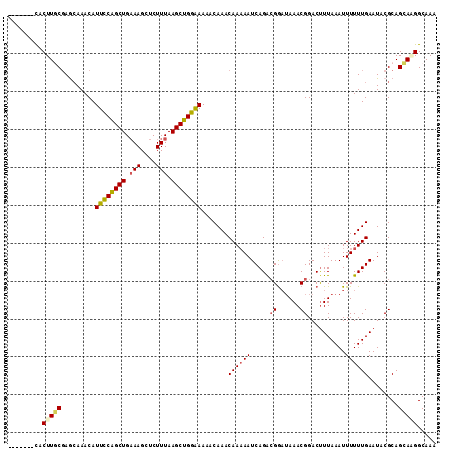
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:37 2006