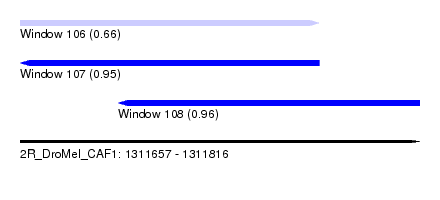
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,311,657 – 1,311,816 |
| Length | 159 |
| Max. P | 0.960433 |

| Location | 1,311,657 – 1,311,776 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.81 |
| Mean single sequence MFE | -46.60 |
| Consensus MFE | -46.31 |
| Energy contribution | -46.62 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.02 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.656958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1311657 119 + 20766785 AUUUGCGGGACACUGAUUCGCCAGU-UUACUUGCGAAGUGUGGGUGCCAAAAGAGGUCCGGCUGUUGGGGUCCUUCAGCAGCCAAACGCUCCGGCACGUGACAUUGGCAAAGUGAGUUCC ......(((((((((......))))-((((((((..(((((.((((((....(((((..((((((((((....))))))))))..)).))).))))).).))))).)).))))))))))) ( -47.00) >DroSec_CAF1 30062 120 + 1 AUUUGCGGGACACUGAUUCGCCAGUUUUACUUGCGAAGUGUGGGUGCCAGAAGAGGUCCGGCUGUUGGGGUCCUUCAGCAGCCAAACGCUCCGGCACGUGACAUUGGCAAAGUGAGUUCC ......(((((((((......)))).((((((((..(((((.((((((....(((((..((((((((((....))))))))))..)).))).))))).).))))).)).))))))))))) ( -47.00) >DroSim_CAF1 9007 120 + 1 AUUUGCGGGACACUGAUUCGCCAGUUUUACUUGCGAAGUGUGGGUGCCAGAAGAGGUCCGGCUGUUGGGGUCCUUCAGCAGCCAAACGCUCCGGCACGUGACAUUGGCAAAGUGAGUUCC ......(((((((((......)))).((((((((..(((((.((((((....(((((..((((((((((....))))))))))..)).))).))))).).))))).)).))))))))))) ( -47.00) >DroEre_CAF1 12548 119 + 1 AUUUGCGGGACACUGAUUCGCCAGUUGUACUUGCGAAGUGUGGGCGCCAUAAGAGGUCCGGCUGUUGGGG-CCUCCAGCAGCCAAACGCUCCGGCACGUGACUUUGGCAAAGUGAGUUCC ......(((((.(...((.(((((...(((.((((.(((((.((.(((......)))))(((((((((..-...)))))))))..))))).).))).)))...))))).))..).))))) ( -45.40) >consensus AUUUGCGGGACACUGAUUCGCCAGUUUUACUUGCGAAGUGUGGGUGCCAGAAGAGGUCCGGCUGUUGGGGUCCUUCAGCAGCCAAACGCUCCGGCACGUGACAUUGGCAAAGUGAGUUCC ......(((((.(...((.((((((.((((.((((.(((((.((.(((......)))))((((((((((....))))))))))..))))).).))).)))).)))))).))..).))))) (-46.31 = -46.62 + 0.31)

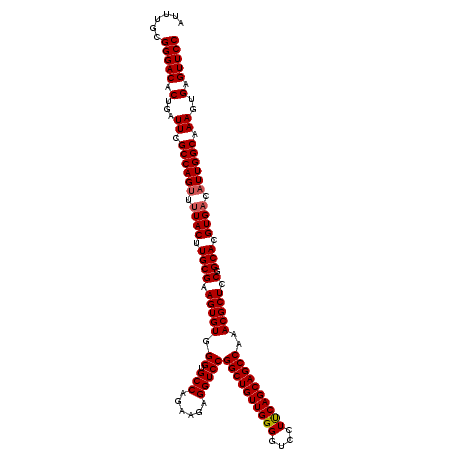
| Location | 1,311,657 – 1,311,776 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.81 |
| Mean single sequence MFE | -42.58 |
| Consensus MFE | -41.94 |
| Energy contribution | -41.75 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953216 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1311657 119 - 20766785 GGAACUCACUUUGCCAAUGUCACGUGCCGGAGCGUUUGGCUGCUGAAGGACCCCAACAGCCGGACCUCUUUUGGCACCCACACUUCGCAAGUAA-ACUGGCGAAUCAGUGUCCCGCAAAU .........(((((.........(((((((((.(((((((((.((........)).)))))))))))))...)))))..(((((((((.((...-.)).))))...)))))...))))). ( -41.50) >DroSec_CAF1 30062 120 - 1 GGAACUCACUUUGCCAAUGUCACGUGCCGGAGCGUUUGGCUGCUGAAGGACCCCAACAGCCGGACCUCUUCUGGCACCCACACUUCGCAAGUAAAACUGGCGAAUCAGUGUCCCGCAAAU .........(((((.........(((((((((.(((((((((.((........)).)))))))))...)))))))))..(((((((((.((.....)).))))...)))))...))))). ( -42.20) >DroSim_CAF1 9007 120 - 1 GGAACUCACUUUGCCAAUGUCACGUGCCGGAGCGUUUGGCUGCUGAAGGACCCCAACAGCCGGACCUCUUCUGGCACCCACACUUCGCAAGUAAAACUGGCGAAUCAGUGUCCCGCAAAU .........(((((.........(((((((((.(((((((((.((........)).)))))))))...)))))))))..(((((((((.((.....)).))))...)))))...))))). ( -42.20) >DroEre_CAF1 12548 119 - 1 GGAACUCACUUUGCCAAAGUCACGUGCCGGAGCGUUUGGCUGCUGGAGG-CCCCAACAGCCGGACCUCUUAUGGCGCCCACACUUCGCAAGUACAACUGGCGAAUCAGUGUCCCGCAAAU .........(((((.........(((((((((.(((((((((.(((...-..))).)))))))))))))...)))))..(((((((((.((.....)).))))...)))))...))))). ( -44.40) >consensus GGAACUCACUUUGCCAAUGUCACGUGCCGGAGCGUUUGGCUGCUGAAGGACCCCAACAGCCGGACCUCUUCUGGCACCCACACUUCGCAAGUAAAACUGGCGAAUCAGUGUCCCGCAAAU .........(((((.........(((((((((.(((((((((.((........)).)))))))))))))...)))))..(((((((((.((.....)).))))...)))))...))))). (-41.94 = -41.75 + -0.19)

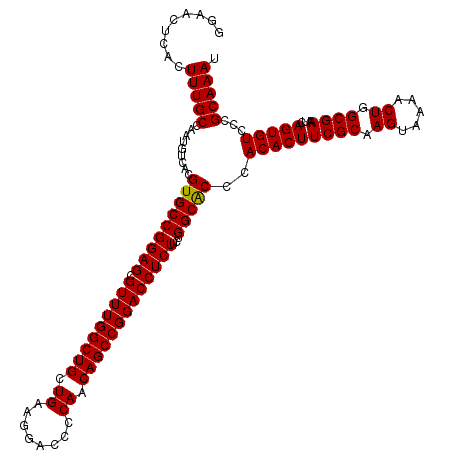
| Location | 1,311,696 – 1,311,816 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -43.38 |
| Consensus MFE | -42.29 |
| Energy contribution | -42.60 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.02 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960433 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1311696 120 - 20766785 CUUGGUCCCAAGAUGACCAAAUUGUGUCACAUUUGCGAACGGAACUCACUUUGCCAAUGUCACGUGCCGGAGCGUUUGGCUGCUGAAGGACCCCAACAGCCGGACCUCUUUUGGCACCCA .((((((.......))))))...(((..(((((.(((((.(.......)))))).))))))))(((((((((.(((((((((.((........)).)))))))))))))...)))))... ( -43.60) >DroSec_CAF1 30102 120 - 1 CUUGGUCCCAAGAUGACCAAAUUGUGUCACAUUUGCGAACGGAACUCACUUUGCCAAUGUCACGUGCCGGAGCGUUUGGCUGCUGAAGGACCCCAACAGCCGGACCUCUUCUGGCACCCA .((((((.......))))))...(((..(((((.(((((.(.......)))))).))))))))(((((((((.(((((((((.((........)).)))))))))...)))))))))... ( -44.30) >DroSim_CAF1 9047 120 - 1 CUUGGUCCCAAGAUGACCAAAUUGUGUCACAUUUGCGAACGGAACUCACUUUGCCAAUGUCACGUGCCGGAGCGUUUGGCUGCUGAAGGACCCCAACAGCCGGACCUCUUCUGGCACCCA .((((((.......))))))...(((..(((((.(((((.(.......)))))).))))))))(((((((((.(((((((((.((........)).)))))))))...)))))))))... ( -44.30) >DroEre_CAF1 12588 119 - 1 CCUGGUCCCAAGAUGACCAAAUUGUGUCACAUUUGCGAACGGAACUCACUUUGCCAAAGUCACGUGCCGGAGCGUUUGGCUGCUGGAGG-CCCCAACAGCCGGACCUCUUAUGGCGCCCA ..(((((.......)))))....(((..((.((.(((((.(.......)))))).)).)))))(((((((((.(((((((((.(((...-..))).)))))))))))))...)))))... ( -41.30) >consensus CUUGGUCCCAAGAUGACCAAAUUGUGUCACAUUUGCGAACGGAACUCACUUUGCCAAUGUCACGUGCCGGAGCGUUUGGCUGCUGAAGGACCCCAACAGCCGGACCUCUUCUGGCACCCA .((((((.......))))))...(((..(((((.(((((.(.......)))))).))))))))(((((((((.(((((((((.((........)).)))))))))))))...)))))... (-42.29 = -42.60 + 0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:25:02 2006