| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,868,577 – 17,868,680 |
| Length | 103 |
| Max. P | 0.993373 |

| Location | 17,868,577 – 17,868,680 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.22 |
| Mean single sequence MFE | -32.67 |
| Consensus MFE | -23.44 |
| Energy contribution | -25.24 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.72 |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.991566 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17868577 103 + 20766785 UGCUCCCCCAACAUC---U-------------C-CUGCUGCAGGACCCAGCAUCCAGGAUCCUGUUUUGUGCAGCGUUUUAAUUGGCUUUUCUGCCAUAUUAGAUUUCAGAGCCUCACUU .((((..........---.-------------.-..((((((.((..(((.(((...))).)))..)).))))))..((((((((((......)))).)))))).....))))....... ( -29.90) >DroSec_CAF1 33442 106 + 1 UGCUCCUCCAGCUCCUCCU-------------C-CAGCUGCAGGACCCAGGAUCCAGGAUCCUGUUUUGUGCAGCGUUUUAAUUGGGUUUUCUGCCAUAUUAGAUUUCAGAGCCUCACUU ..........((((.....-------------.-..((((((.((..(((((((...)))))))..)).))))))..((((((..(((.....)))..)))))).....))))....... ( -33.60) >DroSim_CAF1 33797 103 + 1 UGCUCCUCCAGCUCC---U-------------C-CAGCUGCAGGACCCAGGAUCCAGGAUCCUGUUUUGUGCAGCGUUUUAAUUGGGUUUUCUGCCAUAUUAGAUUUCAGAGCCUCACUU ..........((((.---.-------------.-..((((((.((..(((((((...)))))))..)).))))))..((((((..(((.....)))..)))))).....))))....... ( -33.60) >DroEre_CAF1 35882 120 + 1 UGCUCCACUACCCACUACCCAGUACCCAUUACCCACGAGCCAGGAUCCAGGAUCCAGGAUCCUGUCUUGUGCAGCGUUUUAAUUGGGUUUUCUGCCAUAUUACAUUUCAGAGCCUCAGUU .((((..............(((.(((((..(((((((((.((((((((.(....).)))))))).))))))..).))......)))))...)))...............))))....... ( -33.85) >DroYak_CAF1 32286 113 + 1 UGCUCCAGGACCCAAGACCCAGGAU-------CUAGGAUCCAGGAUCCAGCAACCAGGAUCCUGUUUUGUGCAGCGUUUUAAUUGGGUUUUCUGCCAUAUUAGAUUUCAGAGCCUCAGUU .((((..((....((((((((((((-------(...)))))(((((((........)))))))....................))))))))...))......(....).))))....... ( -32.40) >consensus UGCUCCACCACCCAC__CU_____________C_CAGCUGCAGGACCCAGGAUCCAGGAUCCUGUUUUGUGCAGCGUUUUAAUUGGGUUUUCUGCCAUAUUAGAUUUCAGAGCCUCACUU .((((...............................((((((.((..(((((((...)))))))..)).))))))..(((((((((........))).)))))).....))))....... (-23.44 = -25.24 + 1.80)

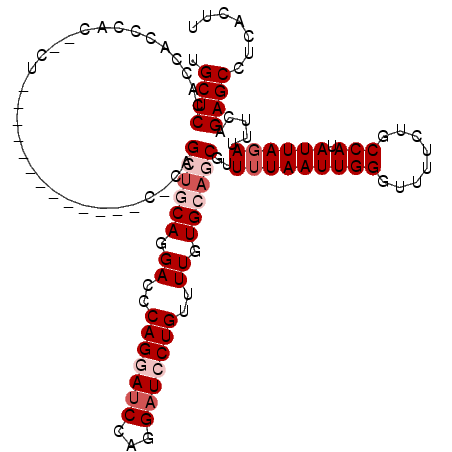
| Location | 17,868,577 – 17,868,680 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.22 |
| Mean single sequence MFE | -36.02 |
| Consensus MFE | -27.92 |
| Energy contribution | -27.08 |
| Covariance contribution | -0.84 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.78 |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.993373 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17868577 103 - 20766785 AAGUGAGGCUCUGAAAUCUAAUAUGGCAGAAAAGCCAAUUAAAACGCUGCACAAAACAGGAUCCUGGAUGCUGGGUCCUGCAGCAG-G-------------A---GAUGUUGGGGGAGCA .......(((((....(((((((((((......))).........((((((.......((((((........))))))))))))..-.-------------.---.))))))))))))). ( -34.91) >DroSec_CAF1 33442 106 - 1 AAGUGAGGCUCUGAAAUCUAAUAUGGCAGAAAACCCAAUUAAAACGCUGCACAAAACAGGAUCCUGGAUCCUGGGUCCUGCAGCUG-G-------------AGGAGGAGCUGGAGGAGCA ..((..((((((....((((...(((........)))........((((((..(..(((((((...)))))))..)..))))))))-)-------------)...))))))......)). ( -34.90) >DroSim_CAF1 33797 103 - 1 AAGUGAGGCUCUGAAAUCUAAUAUGGCAGAAAACCCAAUUAAAACGCUGCACAAAACAGGAUCCUGGAUCCUGGGUCCUGCAGCUG-G-------------A---GGAGCUGGAGGAGCA ..(((.(((((((..((.....))..))))...............))).)))....(((((((...)))))))(.((((.(((((.-.-------------.---..))))).)))).). ( -32.96) >DroEre_CAF1 35882 120 - 1 AACUGAGGCUCUGAAAUGUAAUAUGGCAGAAAACCCAAUUAAAACGCUGCACAAGACAGGAUCCUGGAUCCUGGAUCCUGGCUCGUGGGUAAUGGGUACUGGGUAGUGGGUAGUGGAGCA .......(((((...........(((........)))((((...((((((...((.((((((((.(....).))))))))((((((.....)))))).))..))))))..))))))))). ( -40.10) >DroYak_CAF1 32286 113 - 1 AACUGAGGCUCUGAAAUCUAAUAUGGCAGAAAACCCAAUUAAAACGCUGCACAAAACAGGAUCCUGGUUGCUGGAUCCUGGAUCCUAG-------AUCCUGGGUCUUGGGUCCUGGAGCA .......((((............((((((.................)))).))...((((((((........((((((.(((((...)-------)))).)))))).)))))))))))). ( -37.23) >consensus AAGUGAGGCUCUGAAAUCUAAUAUGGCAGAAAACCCAAUUAAAACGCUGCACAAAACAGGAUCCUGGAUCCUGGGUCCUGCAGCUG_G_____________AG__GGAGCUGGAGGAGCA .......(((((...........((((((.................)))).))...((((((((........))))))))(((((......................)))))..))))). (-27.92 = -27.08 + -0.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:33 2006