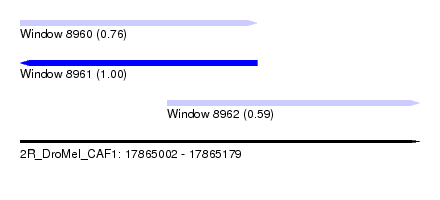
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,865,002 – 17,865,179 |
| Length | 177 |
| Max. P | 0.995702 |

| Location | 17,865,002 – 17,865,107 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 91.55 |
| Mean single sequence MFE | -25.26 |
| Consensus MFE | -21.20 |
| Energy contribution | -21.28 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.757128 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17865002 105 + 20766785 UGUGUGCAUAUGAGCAAAUAUCUCAUUGCAAAAUAUUUGCGUGUACUUCUUAUACAAU-------UCUUGUUAUUGAAUUCGAUCAGAGUUCAAGGAUGGCCAGAUGCGAAU ....((((.(((((.......)))))))))....(((((((((((.....)))))...-------(((.((((((((((((.....)))))))...))))).))).)))))) ( -25.90) >DroSec_CAF1 30326 103 + 1 U--GUGCAUAUGAGCAAAUAUCUCAUGGCAAAAUAUUUGCGUGUACUUCUUAUAAAAU-------UCUUGUUAUUGAAUUCGAUCGGAGUUCAAGGAUGGCCAGAUGCGAAU .--.(((.((((((.......)))))))))....((((((((.(..............-------........((((((((.....))))))))((....))).)))))))) ( -22.70) >DroSim_CAF1 30660 103 + 1 U--GUGCAUAUGAGCAAAUAUCUCAUGGCAAAAUAUUUGCGUGUACUUCUUAUACAAU-------UCUUGUUAUUGAAUUCGAUCGGAGUUCAAGGAUGGCCAGAUGCGAAU .--.(((.((((((.......)))))))))....(((((((((((.....)))))...-------(((.((((((((((((.....)))))))...))))).))).)))))) ( -25.40) >DroEre_CAF1 32829 102 + 1 U--GUGCAUAUGAGCAAAUAUCUCAUUGCAAAAUAUUUGCGUGUCCUUCUUGUACGUU-------UCUUGUUAUUGAAUUCGAUCGGAGUUCAAGGAUGGCCAGAUGCGAA- .--(((((.(((((.......))))).((((.....))))..........)))))((.-------(((.((((((((((((.....)))))))...))))).))).))...- ( -23.00) >DroYak_CAF1 29009 109 + 1 U--GUGCAUAUGAGCAAAUAUCUCAUUGCAAAAUAUUUGCGUGUACUUCUUAUACGAAACGUAUAAGUUGUUAUUGAAUUCGAUCGGAGUUCAAGAAUGGCCAGAUGCGAA- .--.((((.(((((.......))))))))).........((((((.....))))))...(((((..((..((.((((((((.....)))))))).))..))...)))))..- ( -29.30) >consensus U__GUGCAUAUGAGCAAAUAUCUCAUUGCAAAAUAUUUGCGUGUACUUCUUAUACAAU_______UCUUGUUAUUGAAUUCGAUCGGAGUUCAAGGAUGGCCAGAUGCGAAU ....(((((.((.((((((((...........))))))))(((((.....)))))............(..((.((((((((.....)))))))).))..).)).)))))... (-21.20 = -21.28 + 0.08)

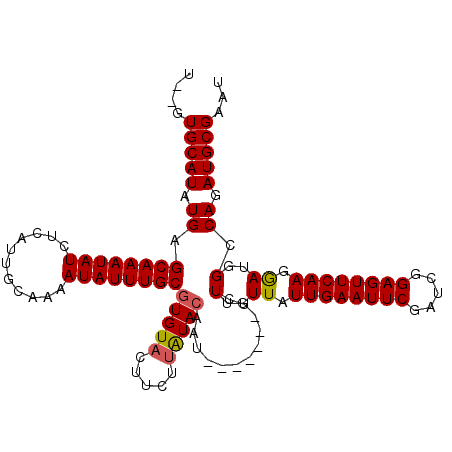
| Location | 17,865,002 – 17,865,107 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 91.55 |
| Mean single sequence MFE | -22.28 |
| Consensus MFE | -19.20 |
| Energy contribution | -19.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.995702 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17865002 105 - 20766785 AUUCGCAUCUGGCCAUCCUUGAACUCUGAUCGAAUUCAAUAACAAGA-------AUUGUAUAAGAAGUACACGCAAAUAUUUUGCAAUGAGAUAUUUGCUCAUAUGCACACA ....((((.(((.....((((.....(((......)))....)))).-------..(((((.....))))).(((((((((((.....)))))))))))))).))))..... ( -21.40) >DroSec_CAF1 30326 103 - 1 AUUCGCAUCUGGCCAUCCUUGAACUCCGAUCGAAUUCAAUAACAAGA-------AUUUUAUAAGAAGUACACGCAAAUAUUUUGCCAUGAGAUAUUUGCUCAUAUGCAC--A ....((((.(((.......((.((((..((.((((((........))-------)))).))..).))).)).(((((((((((.....)))))))))))))).))))..--. ( -21.70) >DroSim_CAF1 30660 103 - 1 AUUCGCAUCUGGCCAUCCUUGAACUCCGAUCGAAUUCAAUAACAAGA-------AUUGUAUAAGAAGUACACGCAAAUAUUUUGCCAUGAGAUAUUUGCUCAUAUGCAC--A ....((((.(((......(((((.((.....)).)))))........-------..(((((.....))))).(((((((((((.....)))))))))))))).))))..--. ( -20.80) >DroEre_CAF1 32829 102 - 1 -UUCGCAUCUGGCCAUCCUUGAACUCCGAUCGAAUUCAAUAACAAGA-------AACGUACAAGAAGGACACGCAAAUAUUUUGCAAUGAGAUAUUUGCUCAUAUGCAC--A -...((((.(((...((((((((.((.....)).)))..........-------..(......))))))...(((((((((((.....)))))))))))))).))))..--. ( -21.00) >DroYak_CAF1 29009 109 - 1 -UUCGCAUCUGGCCAUUCUUGAACUCCGAUCGAAUUCAAUAACAACUUAUACGUUUCGUAUAAGAAGUACACGCAAAUAUUUUGCAAUGAGAUAUUUGCUCAUAUGCAC--A -...((((.(((......(((((.((.....)).)))))..((..((((((((...))))))))..))....(((((((((((.....)))))))))))))).))))..--. ( -26.50) >consensus AUUCGCAUCUGGCCAUCCUUGAACUCCGAUCGAAUUCAAUAACAAGA_______AUUGUAUAAGAAGUACACGCAAAUAUUUUGCAAUGAGAUAUUUGCUCAUAUGCAC__A ....((((.(((......(((((.((.....)).))))).................................(((((((((((.....)))))))))))))).))))..... (-19.20 = -19.20 + -0.00)

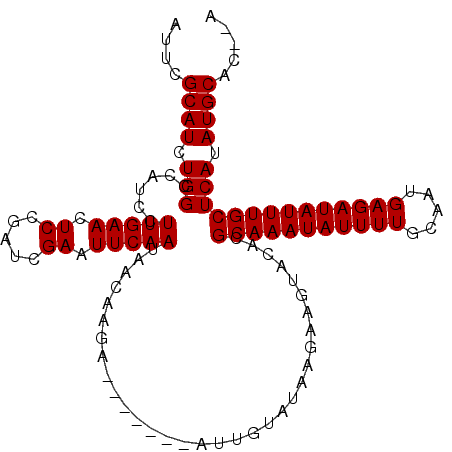

| Location | 17,865,067 – 17,865,179 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 96.99 |
| Mean single sequence MFE | -25.60 |
| Consensus MFE | -23.88 |
| Energy contribution | -23.92 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.593514 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17865067 112 + 20766785 AUUGAAUUCGAUCAGAGUUCAAGGAUGGCCAGAUGCGAAUAUGCCA---AAUUGAAAAUGCAUUGAAUUUUAAGCCGAUUGGGGUCCAAAGAAACCACUCAACCUAAAUCAGCUU ...(((((((((((...(((((...((((...((....))..))))---..)))))..)).))))))))).((((.((((.((((....((......))..)))).)))).)))) ( -26.30) >DroSec_CAF1 30389 112 + 1 AUUGAAUUCGAUCGGAGUUCAAGGAUGGCCAGAUGCGAAUAUGCCA---AAUUGAAAAUGCAUUGAAUUUUAAGCCGAUUGGGGUCUAAAGAAACCACUCAACCUAAAUCAGCUU ...(((((((((((...(((((...((((...((....))..))))---..)))))..)).))))))))).((((.((((.((((....((......))..)))).)))).)))) ( -25.60) >DroSim_CAF1 30723 112 + 1 AUUGAAUUCGAUCGGAGUUCAAGGAUGGCCAGAUGCGAAUAUGCCA---AAUUGAAAAUGCAUUGAAUUUUAAGCCGAUUGGGGUCCAAAGAAACCACUCAACCUAAAUCAGCUU ...(((((((((((...(((((...((((...((....))..))))---..)))))..)).))))))))).((((.((((.((((....((......))..)))).)))).)))) ( -25.60) >DroEre_CAF1 32892 111 + 1 AUUGAAUUCGAUCGGAGUUCAAGGAUGGCCAGAUGCGAA-AUGCCA---AAUUGAAAAUGCAUUGAAAUUUAAGCCGAUUGGGGUCCAAAGAAACCACUCAACCUAAAUCAGCUU ..(((((((.....)))))))(((..(((.((((....(-((((..---..........)))))...))))..)))((.(((..((....))..))).))..))).......... ( -24.20) >DroYak_CAF1 29079 114 + 1 AUUGAAUUCGAUCGGAGUUCAAGAAUGGCCAGAUGCGAA-AUGCCAUUGAAUUGAAAAUGCAUUGAAUUUUAAGCCGAUUGGGGUCCAAAGAAACCACUCAACCUAAAUCAGCUU ...(((((((((((...(((((.((((((..........-..))))))...)))))..)).))))))))).((((.((((.((((....((......))..)))).)))).)))) ( -26.30) >consensus AUUGAAUUCGAUCGGAGUUCAAGGAUGGCCAGAUGCGAAUAUGCCA___AAUUGAAAAUGCAUUGAAUUUUAAGCCGAUUGGGGUCCAAAGAAACCACUCAACCUAAAUCAGCUU ...(((((((((((...(((((...((((.............)))).....)))))..)).))))))))).((((.((((.((((....((......))..)))).)))).)))) (-23.88 = -23.92 + 0.04)

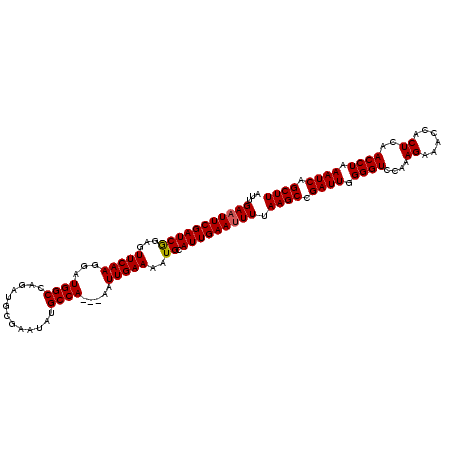
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:29 2006