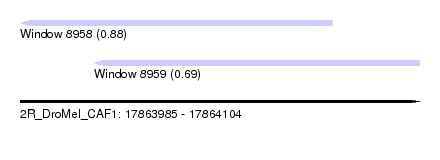
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,863,985 – 17,864,104 |
| Length | 119 |
| Max. P | 0.884536 |

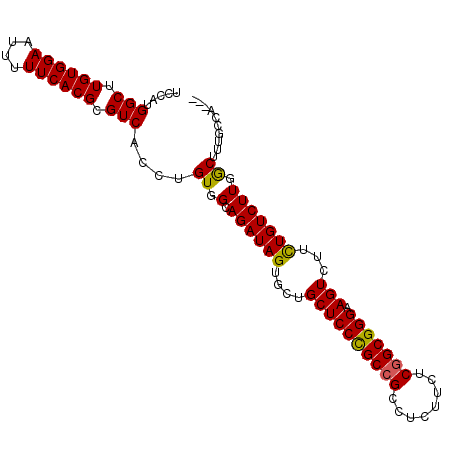
| Location | 17,863,985 – 17,864,078 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 80.57 |
| Mean single sequence MFE | -36.75 |
| Consensus MFE | -27.05 |
| Energy contribution | -26.05 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.884536 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17863985 93 - 20766785 GUCACCUGUGGCAGAUAGUGCUGCUCCCGCCGCCUCUUCUCGGCGGGAAGUCUUCUGUCUUGACUUUGUCG---AGGGAGUCGGAGUUUUGCCAUA ......(((((((((.....((((((((.(((((.......)))))((((((.........))))))....---.))))).)))...))))))))) ( -38.60) >DroSec_CAF1 29299 93 - 1 GUCACCUGUGGCAGAUAGUGGUGCUCCCGCCGCCUCUUCUCGGCGGGAAGUCUUCUGUCUUGGCUUUGUCG---AGGGAGUCGUAGUACUGUCAUA ((((....)))).(((((((.(((((((((((........))))))))......((.(((((((...))))---))).))..))).)))))))... ( -40.10) >DroEre_CAF1 31825 86 - 1 GUCACCUGUGGCAGAUAGUGCAGCUCCCGCCGCCUCUUCUCGGCGGGAAGUCUGUUGUCUUGGCUUGGCCA---GGGGGGUAAGAGG-GU------ ....(((.(((((((((..(((((((((((((........))))))).)).))))))))).......))))---.))).........-..------ ( -34.41) >DroYak_CAF1 27967 95 - 1 GUCACCUGUGGCAGAUAGUGGUGCUCCUGCUGCCUAUUCUCUGCGGGAAGUCUUCUGUCUUGGCUUUACAAAAUGGGGGGCAAGAGG-GUGCCAUA ((((....)))).....((((..((((((((.((((((...((...((((((.........)))))).)).)))))).))).)).))-)..)))). ( -33.90) >consensus GUCACCUGUGGCAGAUAGUGCUGCUCCCGCCGCCUCUUCUCGGCGGGAAGUCUUCUGUCUUGGCUUUGCCA___AGGGAGUAAGAGG_GUGCCAUA ((((....))))((((((....((((((((((........))))))).)))...))))))((((((..(.....)..))))))............. (-27.05 = -26.05 + -1.00)

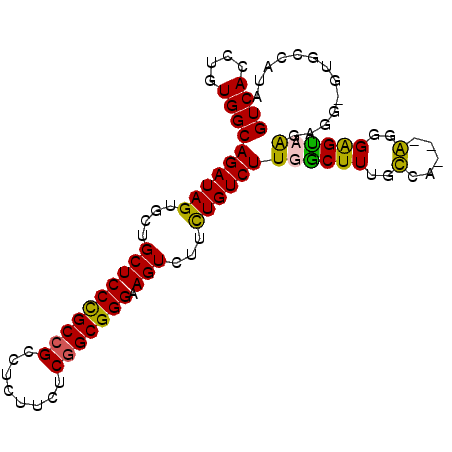
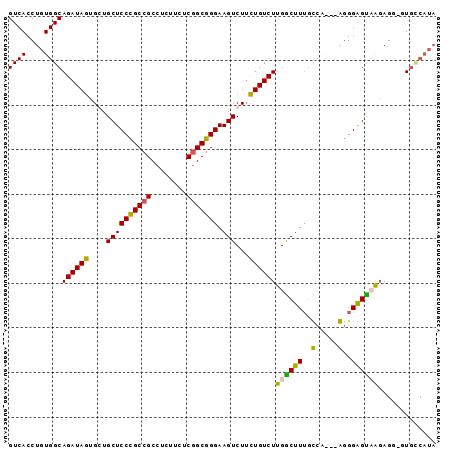
| Location | 17,864,007 – 17,864,104 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 87.88 |
| Mean single sequence MFE | -34.57 |
| Consensus MFE | -28.59 |
| Energy contribution | -28.27 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.685221 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17864007 97 - 20766785 UCCAUGGCUUGUGGAAUUUUUCACGCGUCACCUGUGGCAGAUAGUGCUGCUCCCGCCGCCUCUUCUCGGCGGGAAGUCUUCUGUCUUGACUUUGUCG--- ....((((.((((((....)))))).)))).....((((((.((.(((..((((((((........)))))))))))))))))))..(((...))).--- ( -34.00) >DroEre_CAF1 31840 97 - 1 UCCAUGGCUUGUGGAAUUUUUCACGCGUCACCUGUGGCAGAUAGUGCAGCUCCCGCCGCCUCUUCUCGGCGGGAAGUCUGUUGUCUUGGCUUGGCCA--- ....(((((.(((((....)))))((((((....))))(((((..(((((((((((((........))))))).)).)))))))))..))..)))))--- ( -38.40) >DroYak_CAF1 27988 100 - 1 UCCAAGGCUUGUGGAAUUUUUCACGCGUCACCUGUGGCAGAUAGUGGUGCUCCUGCUGCCUAUUCUCUGCGGGAAGUCUUCUGUCUUGGCUUUACAAAAU .(((((((....(((.(((..(.((((.....))))(((((.(((((.((.......))))))).))))))..))))))...)))))))........... ( -31.30) >consensus UCCAUGGCUUGUGGAAUUUUUCACGCGUCACCUGUGGCAGAUAGUGCUGCUCCCGCCGCCUCUUCUCGGCGGGAAGUCUUCUGUCUUGGCUUUGCCA___ .....(((.((((((....)))))).)))....((.(.((((((....((((((((((........))))))).)))...))))))).)).......... (-28.59 = -28.27 + -0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:26 2006