| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,861,479 – 17,861,706 |
| Length | 227 |
| Max. P | 0.869037 |

| Location | 17,861,479 – 17,861,587 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 81.17 |
| Mean single sequence MFE | -27.47 |
| Consensus MFE | -19.07 |
| Energy contribution | -20.17 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866633 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17861479 108 + 20766785 AUAUGUCAGAAGUAAGCCUAUUUCGUUUACGAAAUGGUUUCUGCAUUUUCUCUCUGUGCAGGUAAAUAUGAUUUUCGUGUCUGGCAGCAAUUACUGAUGUUGUCAUUA ...(((((((...(((((.((((((....)))))))))))((((((.........)))))).....((((.....)))))))))))(((((.......)))))..... ( -27.50) >DroSec_CAF1 26803 108 + 1 AUAUGUCAGAAGUAAGCCUAUUUUGCUUACGAAAUGGUUUCUGCAUUUUCUCUCUGUGCAGGUAAAUAUGACUGUCGUGUCUGGCAACAAUUACUGGUGUUGUCAUUA (((((.(((..((((((.......)))))).........(((((((.........))))))).........))).))))).((((((((........))))))))... ( -29.00) >DroEre_CAF1 29424 93 + 1 UCAUAGCAGAAG-------------CUUAUGAAUUGGCUUCUGCAUUUUCUCUCUGUGCAGUUAAG--UGCCUGUCGUGUCUGGCAACAAUUACUGGUGUUGUCAUUA .(((.(((((((-------------((........))))))))).............((((.....--...)))).)))..((((((((........))))))))... ( -25.90) >consensus AUAUGUCAGAAGUAAGCCUAUUU_GCUUACGAAAUGGUUUCUGCAUUUUCUCUCUGUGCAGGUAAAUAUGACUGUCGUGUCUGGCAACAAUUACUGGUGUUGUCAUUA .((((.(((..((((((.......))))))..........((((((.........))))))..........))).))))..((((((((........))))))))... (-19.07 = -20.17 + 1.09)

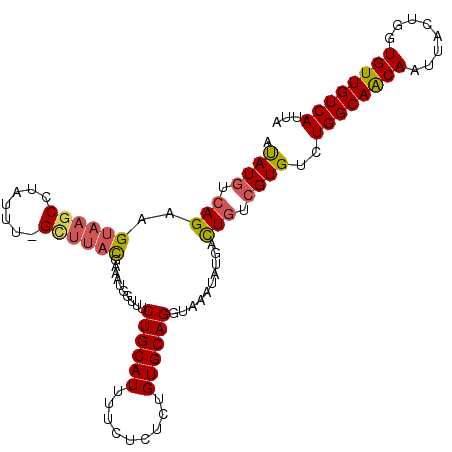
| Location | 17,861,551 – 17,861,666 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 95.97 |
| Mean single sequence MFE | -29.17 |
| Consensus MFE | -26.14 |
| Energy contribution | -25.70 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.869037 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17861551 115 - 20766785 UAAUCAAAGUGUUUGCACGCAUUUUUAAACCGAUUUGCGCCUGUGCAAAUCUUUAUUAAUUUCCGAGUG-GUGUUCUUACUAAUGACAACAUCAGUAAUUGCUGCCAGACACGAAA ........(((((((((.(((..........((((((((....))))))))..............(.((-(((((.(((....))).))))))).)...))))).))))))).... ( -29.50) >DroSec_CAF1 26875 115 - 1 UAAUCAAAGUGUUUGCACGCAUUUUUAAACCGAUUUGCGCCUGUGCAAAUCUUUAUUAAUUUCCGAGUG-GUGUUCUUACUAAUGACAACACCAGUAAUUGUUGCCAGACACGACA ........(((((((((.(((..........((((((((....))))))))..............(.((-(((((.(((....))).))))))).)...))))).))))))).... ( -29.70) >DroEre_CAF1 29481 116 - 1 UUAUCAAAGUGUUUGCACGCAUUUUUAAACCGAUUUGCGCCUGUGCAAAUCCUUACUAAUUUCCGAGUGGGUGUUCUUACUAAUGACAACACCAGUAAUUGUUGCCAGACACGACA ........(((((((((.(((..........((((((((....))))))))..((((........))))((((((.(((....))).))))))......))))).))))))).... ( -28.30) >consensus UAAUCAAAGUGUUUGCACGCAUUUUUAAACCGAUUUGCGCCUGUGCAAAUCUUUAUUAAUUUCCGAGUG_GUGUUCUUACUAAUGACAACACCAGUAAUUGUUGCCAGACACGACA ........(((((((((.(((..........((((((((....))))))))..............(.((.(((((.(((....))).))))))).)...))))).))))))).... (-26.14 = -25.70 + -0.44)

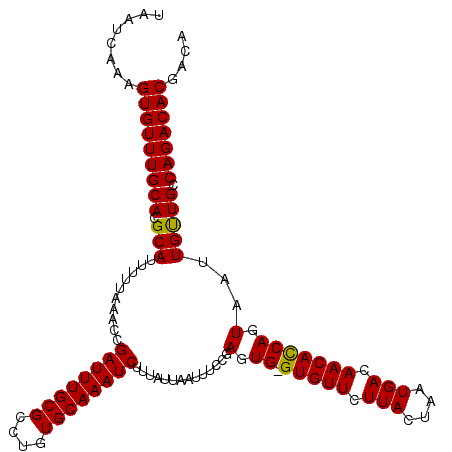
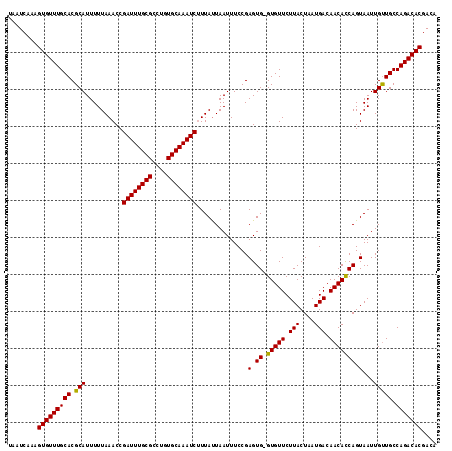
| Location | 17,861,587 – 17,861,706 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.66 |
| Mean single sequence MFE | -33.43 |
| Consensus MFE | -30.60 |
| Energy contribution | -30.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784105 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17861587 119 - 20766785 UCAUUUGGGAAUUUCGGGCCGCUUGGGUUUUGUGAAUGAAUAAUCAAAGUGUUUGCACGCAUUUUUAAACCGAUUUGCGCCUGUGCAAAUCUUUAUUAAUUUCCGAGUG-GUGUUCUUAC .....(((((((.....(((((((((((((...(((((..........(((....))).)))))..)))))((((((((....))))))))............))))))-))))))))). ( -34.20) >DroSec_CAF1 26911 119 - 1 UCAUUUGGGAAUUUCGGGCCGCUUGGGUUUUGUGAAUGAAUAAUCAAAGUGUUUGCACGCAUUUUUAAACCGAUUUGCGCCUGUGCAAAUCUUUAUUAAUUUCCGAGUG-GUGUUCUUAC .....(((((((.....(((((((((((((...(((((..........(((....))).)))))..)))))((((((((....))))))))............))))))-))))))))). ( -34.20) >DroEre_CAF1 29517 120 - 1 UCAUUUGGGAAUUUCUGGCCGCCUGGGUUUUGUGAAUGAAUUAUCAAAGUGUUUGCACGCAUUUUUAAACCGAUUUGCGCCUGUGCAAAUCCUUACUAAUUUCCGAGUGGGUGUUCUUAC (((((((((((((...((.......(((((...(((((..........(((....))).)))))..)))))((((((((....))))))))))....))))))))))))).......... ( -31.90) >consensus UCAUUUGGGAAUUUCGGGCCGCUUGGGUUUUGUGAAUGAAUAAUCAAAGUGUUUGCACGCAUUUUUAAACCGAUUUGCGCCUGUGCAAAUCUUUAUUAAUUUCCGAGUG_GUGUUCUUAC .((((((((((((............(((((...(((((..........(((....))).)))))..)))))((((((((....))))))))......))))))))))))........... (-30.60 = -30.60 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:20 2006