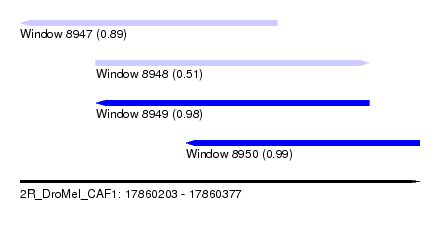
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,860,203 – 17,860,377 |
| Length | 174 |
| Max. P | 0.989457 |

| Location | 17,860,203 – 17,860,315 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.07 |
| Mean single sequence MFE | -37.10 |
| Consensus MFE | -27.18 |
| Energy contribution | -27.26 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.889044 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17860203 112 - 20766785 GCAAAGCUGAUUGCUGUCAGUUUUGCGGCCGAAGGAGGAGCAGCAUUCUACUGGCGAUG-UAGAUUGCAGCGAAAUCGCUUGGCAAA--GAACAGGGGCGGAGAUUGGU-----UUAUGU (((((((..((..(((((.(((((...(((((..((...((.(((.(((((.......)-)))).))).))....))..)))))..)--))))...)))))..))..))-----)).))) ( -39.90) >DroSec_CAF1 25587 110 - 1 GCAAAGCUGAUUGCUGUCAGUUUUGCGGCCGAAGGAGGA-CAGCAUUCUGCUGGCGAUG-UAGAUUGCAGCGAAAUCGUUUGGCAAA--CAACAGGGGCGGAGAU-GGU-----UUAUGU (((((((((((....))))))))))).((((((.((...-((((.....))))(((((.-...))))).......)).))))))...--..(((..(((......-.))-----)..))) ( -38.60) >DroSim_CAF1 26266 111 - 1 GCAAAGCUGAUUGCUGUCAGUUUUGCGGCCGAAGGAGGAGCAGCAUUCUGCUGGCGAUG-UAGAUUGCAGAGAAAUCGCUUGGCAAA--CAACAGGGGCGGAGAU-GGU-----UUAUGU (((((((((((....))))))))))).(((.......((((.(((.(((((.......)-)))).))).((....)))))))))...--..(((..(((......-.))-----)..))) ( -37.21) >DroEre_CAF1 28237 112 - 1 GCAAAGCUGAUUGCUGUCAGUUUUGCGGCCGAAGAAGGAGAA------UGCUGGCGAUG-UAGAUUGCAGCGAAAUCGCUUGGCAAACACAACAGGGGCGGAGAU-AGGGGGGCUUAUAA (((((((((((....)))))))))))((((.......((...------(((((.((((.-...)))))))))...)).(((.((...(......)..)).)))..-.....))))..... ( -35.50) >DroYak_CAF1 25926 108 - 1 GCAAAGCUGAUUGCUGUCAGUUUUGCGGCAAAAGGCGGAGAA------UGCUGGCGAUGGUAGAUUGCAGCGAAAUCGCUUGGCAAACACAACAGGAGCGGCGAU-GAUG-----CAUAA (((((((((((....))))))))))).((...((((((....------(((((.((((.....)))))))))...)))))).)).............(((.....-..))-----).... ( -34.30) >consensus GCAAAGCUGAUUGCUGUCAGUUUUGCGGCCGAAGGAGGAGCAGCAUUCUGCUGGCGAUG_UAGAUUGCAGCGAAAUCGCUUGGCAAA__CAACAGGGGCGGAGAU_GGU_____UUAUGU (((((((((((....))))))))))).(((((.....................(((((.....))))).(((....)))))))).................................... (-27.18 = -27.26 + 0.08)

| Location | 17,860,236 – 17,860,355 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.43 |
| Mean single sequence MFE | -30.67 |
| Consensus MFE | -24.16 |
| Energy contribution | -25.22 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.79 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.511137 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17860236 119 + 20766785 AGCGAUUUCGCUGCAAUCUA-CAUCGCCAGUAGAAUGCUGCUCCUCCUUCGGCCGCAAAACUGACAGCAAUCAGCUUUGCUGUGGAUAAAGUUUUAUGCCGAGAAACUCAUUUAGUUUGC ...((((((((.(((.((((-(.......))))).))).)).......(((((...((((((.(((((((......)))))))......))))))..))))))))).))........... ( -34.60) >DroSec_CAF1 25619 118 + 1 AACGAUUUCGCUGCAAUCUA-CAUCGCCAGCAGAAUGCUG-UCCUCCUUCGGCCGCAAAACUGACAGCAAUCAGCUUUGCUGUGGAUAAAGUUUUAUGCCGAGAAACUCAUUUAGUUUGC ............((((.(((-......((((.....))))-......((((((...((((((.(((((((......)))))))......))))))..)))))).........))).)))) ( -29.10) >DroSim_CAF1 26298 119 + 1 AGCGAUUUCUCUGCAAUCUA-CAUCGCCAGCAGAAUGCUGCUCCUCCUUCGGCCGCAAAACUGACAGCAAUCAGCUUUGCUGUGGAUAAAGUUUUAUGCCGAGAAACUCAUUUAGUUUGC ...((((((((.((((((((-((..((((((((....)))))........))).(((((.((((......)))).)))))))))))).........))).)))))).))........... ( -33.20) >DroEre_CAF1 28276 113 + 1 AGCGAUUUCGCUGCAAUCUA-CAUCGCCAGCA------UUCUCCUUCUUCGGCCGCAAAACUGACAGCAAUCAGCUUUGCUGUGGAUAAAGUUUUAUGCCGAGAAACUCAUUUAGUUUGC ((((....))))((((.(((-........(((------(....(((..((.((.(((((.((((......)))).))))).)).))..)))....)))).(((...)))...))).)))) ( -29.90) >DroYak_CAF1 25960 114 + 1 AGCGAUUUCGCUGCAAUCUACCAUCGCCAGCA------UUCUCCGCCUUUUGCCGCAAAACUGACAGCAAUCAGCUUUGCUGUGGAUAAAGUUUUAUGCCGAGAAACUCAUUUAGUUUGC ((((....))))((((.(((.........(((------(.........((..(.(((((.((((......)))).))))).)..)).........)))).(((...)))...))).)))) ( -26.57) >consensus AGCGAUUUCGCUGCAAUCUA_CAUCGCCAGCAGAAUGCUGCUCCUCCUUCGGCCGCAAAACUGACAGCAAUCAGCUUUGCUGUGGAUAAAGUUUUAUGCCGAGAAACUCAUUUAGUUUGC ((((....))))((((.(((........(((((....))))).....((((((...((((((.(((((((......)))))))......))))))..)))))).........))).)))) (-24.16 = -25.22 + 1.06)

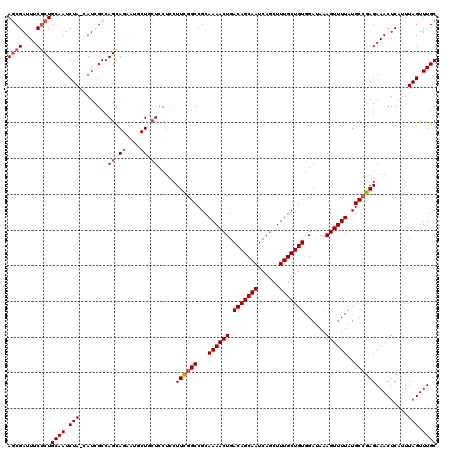
| Location | 17,860,236 – 17,860,355 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.43 |
| Mean single sequence MFE | -38.86 |
| Consensus MFE | -30.33 |
| Energy contribution | -30.97 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.976965 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17860236 119 - 20766785 GCAAACUAAAUGAGUUUCUCGGCAUAAAACUUUAUCCACAGCAAAGCUGAUUGCUGUCAGUUUUGCGGCCGAAGGAGGAGCAGCAUUCUACUGGCGAUG-UAGAUUGCAGCGAAAUCGCU .............(((((((((.((((....))))))...(((((((((((....)))))))))))........))))))).(((.(((((.......)-)))).)))((((....)))) ( -41.30) >DroSec_CAF1 25619 118 - 1 GCAAACUAAAUGAGUUUCUCGGCAUAAAACUUUAUCCACAGCAAAGCUGAUUGCUGUCAGUUUUGCGGCCGAAGGAGGA-CAGCAUUCUGCUGGCGAUG-UAGAUUGCAGCGAAAUCGUU ((((.(((.....((((((((((((((....)))).....(((((((((((....))))))))))).)))).))))...-((((.....))))))....-))).))))((((....)))) ( -37.90) >DroSim_CAF1 26298 119 - 1 GCAAACUAAAUGAGUUUCUCGGCAUAAAACUUUAUCCACAGCAAAGCUGAUUGCUGUCAGUUUUGCGGCCGAAGGAGGAGCAGCAUUCUGCUGGCGAUG-UAGAUUGCAGAGAAAUCGCU ((...........(((((((((.((((....))))))...(((((((((((....)))))))))))........))))))).(((.(((((.......)-)))).))).((....)))). ( -40.50) >DroEre_CAF1 28276 113 - 1 GCAAACUAAAUGAGUUUCUCGGCAUAAAACUUUAUCCACAGCAAAGCUGAUUGCUGUCAGUUUUGCGGCCGAAGAAGGAGAA------UGCUGGCGAUG-UAGAUUGCAGCGAAAUCGCU ((((.(((.....(((.(.((((((....((((.((....(((((((((((....)))))))))))....)).))))....)------)))))).))).-))).))))((((....)))) ( -38.10) >DroYak_CAF1 25960 114 - 1 GCAAACUAAAUGAGUUUCUCGGCAUAAAACUUUAUCCACAGCAAAGCUGAUUGCUGUCAGUUUUGCGGCAAAAGGCGGAGAA------UGCUGGCGAUGGUAGAUUGCAGCGAAAUCGCU ((((.(((.....(((.(.((((((.........(((...(((((((((((....))))))))))).((.....)))))..)------)))))).)))..))).))))((((....)))) ( -36.50) >consensus GCAAACUAAAUGAGUUUCUCGGCAUAAAACUUUAUCCACAGCAAAGCUGAUUGCUGUCAGUUUUGCGGCCGAAGGAGGAGCAGCAUUCUGCUGGCGAUG_UAGAUUGCAGCGAAAUCGCU ((((.(((.....(((.(.(((((..........(((...(((((((((((....)))))))))))...(....).))).........)))))).)))..))).))))((((....)))) (-30.33 = -30.97 + 0.64)

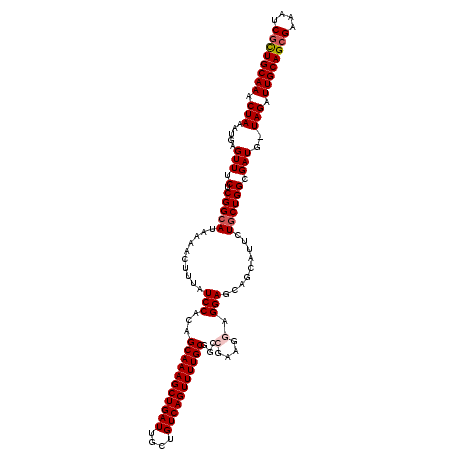
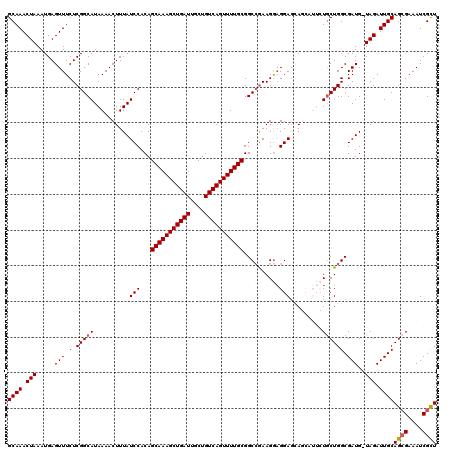
| Location | 17,860,275 – 17,860,377 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 95.69 |
| Mean single sequence MFE | -28.34 |
| Consensus MFE | -25.90 |
| Energy contribution | -26.50 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.989457 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17860275 102 - 20766785 UGCCGGCUGCCACUAAUUAAUUGCAAACUAAAUGAGUUUCUCGGCAUAAAACUUUAUCCACAGCAAAGCUGAUUGCUGUCAGUUUUGCGGCCGAAGGAGGAG ...((((((((...((((.(((........))).))))....))).................(((((((((((....))))))))))))))))......... ( -28.70) >DroSec_CAF1 25658 101 - 1 UGCCGGCUGCCACUAAUUAAUUGCAAACUAAAUGAGUUUCUCGGCAUAAAACUUUAUCCACAGCAAAGCUGAUUGCUGUCAGUUUUGCGGCCGAAGGAGGA- ...((((((((...((((.(((........))).))))....))).................(((((((((((....))))))))))))))))........- ( -28.70) >DroSim_CAF1 26337 102 - 1 UGCCGGCUGCCACUAAUUAAUUGCAAACUAAAUGAGUUUCUCGGCAUAAAACUUUAUCCACAGCAAAGCUGAUUGCUGUCAGUUUUGCGGCCGAAGGAGGAG ...((((((((...((((.(((........))).))))....))).................(((((((((((....))))))))))))))))......... ( -28.70) >DroEre_CAF1 28309 102 - 1 UGCCGGCUGCCACUAAUUAAUUGCAAACUAAAUGAGUUUCUCGGCAUAAAACUUUAUCCACAGCAAAGCUGAUUGCUGUCAGUUUUGCGGCCGAAGAAGGAG ...((((((((...((((.(((........))).))))....))).................(((((((((((....))))))))))))))))......... ( -28.70) >DroYak_CAF1 25994 101 - 1 -NNNNNCUGCCACUAAUUAAUUGCAAACUAAAUGAGUUUCUCGGCAUAAAACUUUAUCCACAGCAAAGCUGAUUGCUGUCAGUUUUGCGGCAAAAGGCGGAG -.....(((((..........((((((((.....)))))....)))................(((((((((((....))))))))))).......))))).. ( -26.90) >consensus UGCCGGCUGCCACUAAUUAAUUGCAAACUAAAUGAGUUUCUCGGCAUAAAACUUUAUCCACAGCAAAGCUGAUUGCUGUCAGUUUUGCGGCCGAAGGAGGAG ...((((((((...((((.(((........))).))))....))).................(((((((((((....))))))))))))))))......... (-25.90 = -26.50 + 0.60)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:17 2006