| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,788,210 – 17,788,302 |
| Length | 92 |
| Max. P | 0.973323 |

| Location | 17,788,210 – 17,788,302 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
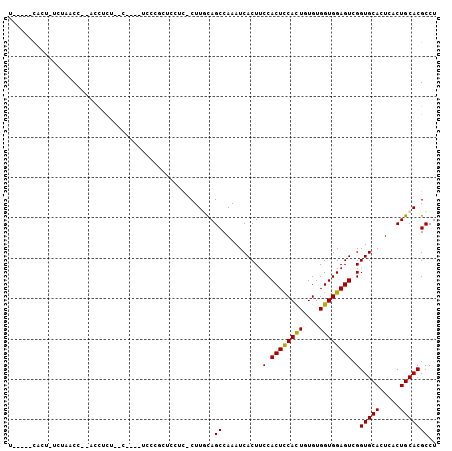
| Mean pairwise identity | 77.51 |
| Mean single sequence MFE | -22.09 |
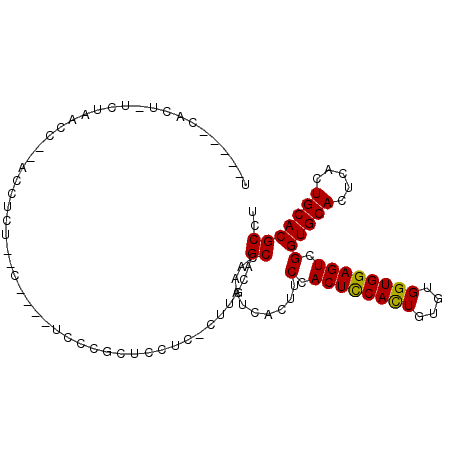
| Consensus MFE | -17.02 |
| Energy contribution | -16.55 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973323 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17788210 92 + 20766785 UA----UAUU-UCUAACC--GCUCCC--C----CCUUGAUUCUC-CUUUCAGCCAAAUCACUUCCACUUCACUGUGUGGUGGAGUCGGUGCACUCACUGCACGCCU ..----....-....(((--(((((.--.----..((((.....-...))))...........((((........)))).)))).))))(((.....)))...... ( -19.20) >DroPse_CAF1 62917 92 + 1 ------CACU-UCUAACGCGACCUCU--C----UCUCGCUCUCU-CUUGCAGCCAAAUCACUUCCACUCCAUUGUGUGGUGGAGUCGGUGCACUCACUGCACGCCU ------....-......((((.....--.----..)))).....-..(((((......((((...((((((((....)))))))).))))......)))))..... ( -22.90) >DroEre_CAF1 57688 95 + 1 UG----CACU-UCUAACC--GCCUCU--GCCGCU-CUCCUCCUC-CUUGCAGCCAAAUCACUUCCACUUCACUGUGUGGUGGAGUCGGUGCACUCCCUGCACGCCU .(----(...-....(((--(.(((.--.((((.-.........-...))..............(((......))).))..))).))))(((.....)))..)).. ( -23.12) >DroYak_CAF1 60013 102 + 1 UAUAUAUAUU-ACUAACC--GCCUCCUUUCUACUCCUUCUGCUC-CUUUUAGCCAAAUCACUUCCACUUCACUGUGUGGUGGAGUCGGUGCACUCCCUGCACGCCU ..........-....(((--(.((((..............(((.-.....)))..........((((........)))).)))).))))(((.....)))...... ( -19.80) >DroAna_CAF1 58083 94 + 1 UC----UGUCCCCUAACC--AGUUUU--C----CUCCCCUCUUCUCUUGCAGCCAAAUCACUUCCACUCCACUGUGUGGUGGAGUCGGUGCACUCGCUGCACGCCU ..----............--......--.----..............((((((.....((((...((((((((....)))))))).)))).....))))))..... ( -24.60) >DroPer_CAF1 63478 92 + 1 ------CACU-UCUAACGCGACCUCU--C----UCUCGCUCUCU-CUUGCAGCCAAAUCACUUCCACUCCAUUGUGUGGUGGAGUCGGUGCACUCACUGCACGCCU ------....-......((((.....--.----..)))).....-..(((((......((((...((((((((....)))))))).))))......)))))..... ( -22.90) >consensus U_____CACU_UCUAACC__ACCUCU__C____UCCCGCUCCUC_CUUGCAGCCAAAUCACUUCCACUCCACUGUGUGGUGGAGUCGGUGCACUCACUGCACGCCU ...................................................((..........(.((((((((....)))))))).)(((((.....))))))).. (-17.02 = -16.55 + -0.47)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:48:49 2006