| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,768,556 – 17,768,660 |
| Length | 104 |
| Max. P | 0.990317 |

| Location | 17,768,556 – 17,768,660 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.48 |
| Mean single sequence MFE | -27.56 |
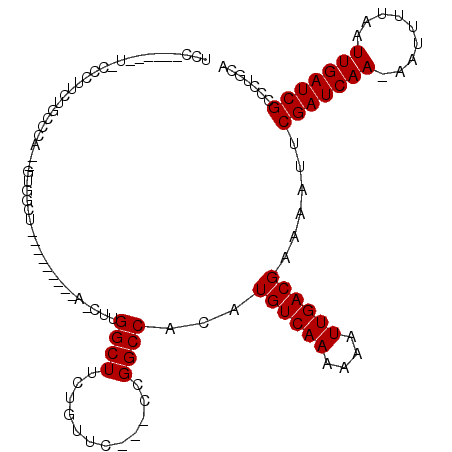
| Consensus MFE | -15.45 |
| Energy contribution | -15.45 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.56 |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.990317 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
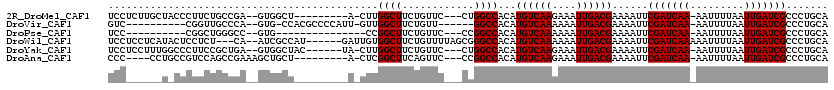
>2R_DroMel_CAF1 17768556 104 - 20766785 UCCUCUUGCUACCCUUCUGCCGA--GUGGCU---------A-CUUGGCUUCUGUUC---CUGGCCACAUGUCAAGAAAUUGACGAAAAUUCGAUCAA-AAUUUUAAUUGAUCGCCCUGCA ......(((.........(((((--((....---------)-))))))........---..(((....((((((....)))))).......((((((-........)))))))))..))) ( -28.00) >DroVir_CAF1 52382 99 - 1 GUC----------CGGUUGCCCA--GUG-CCACGCCCCAUU-GUUGGCUUCUGUU------GGCCACAUGUCAAAAAAUUGACGAAAAUUCGAUCAA-AAUUUUAAUUGAUCGCCCUGCA ((.----------.(((.(((((--(.(-(((.((......-))))))..)))..------)))....((((((....)))))).......((((((-........)))))))))..)). ( -24.10) >DroPse_CAF1 41320 89 - 1 UCC----------CGGCUGGGCC--GUG---------------CCGGCUUCUGUUC---CCGGCCACAUGUCAAAAAAUUGACGAAAAUUCGAUCAA-AAUUUUAAUUGAUCGCCCUGCA ...----------..((.((((.--(((---------------((((.........---)))).))).((((((....)))))).......((((((-........)))))))))).)). ( -29.90) >DroWil_CAF1 56431 109 - 1 UCCUCCUCAUACUCCUCU---CA--AUCGCCAU------GAUUGUGGCUUCUGUUUUAGCGGGCCACAUGUCAAAAAAUUGACGAAAAUUCGAUCAAAAAUUUUAAUUGAUCGCCCUGCA ..................---..--...(((((------....)))))..........((((((....((((((....)))))).......((((((.........)))))))))).)). ( -26.30) >DroYak_CAF1 39245 107 - 1 UCCUCCUUUGGCCCUUCCGCUGA--GUGGCUAC------UA-CUUGGCUUCUGUUC---CUGGCCACAUGUCAAGAAAUUGACGAAAAUUCGAUCAA-AAUUUUAAUUGAUCGCCCUGCA ........(((((.....((..(--((((...)------))-))..))........---..)))))..((((((....))))))......(((((((-........)))))))....... ( -29.06) >DroAna_CAF1 37435 102 - 1 CCC----CCUGCCGUCCAGCCGAAAGCUGCU---------A-CUCGGCUUCAGUUC---CCGGCCACAUGUCAAGAAAUUGACGAAAAUUCGAUCAA-AAUUUUAAUUGAUCGCCCUGCA ...----...((((..(((.(....))))..---------.-..))))....((..---..(((....((((((....)))))).......((((((-........)))))))))..)). ( -28.00) >consensus UCC______U_CCCUUCUGCCCA__GUGGCU_________A_CUUGGCUUCUGUUC___CCGGCCACAUGUCAAAAAAUUGACGAAAAUUCGAUCAA_AAUUUUAAUUGAUCGCCCUGCA .............................................((((............))))...((((((....))))))......(((((((.........)))))))....... (-15.45 = -15.45 + 0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:48:41 2006