| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,749,726 – 17,749,817 |
| Length | 91 |
| Max. P | 0.996341 |

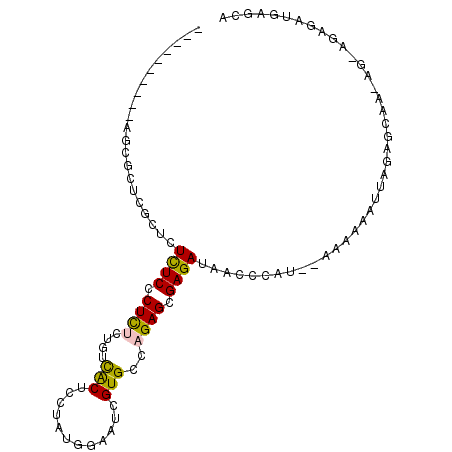
| Location | 17,749,726 – 17,749,817 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 74.71 |
| Mean single sequence MFE | -32.72 |
| Consensus MFE | -16.28 |
| Energy contribution | -16.70 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.50 |
| SVM decision value | 2.68 |
| SVM RNA-class probability | 0.996341 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
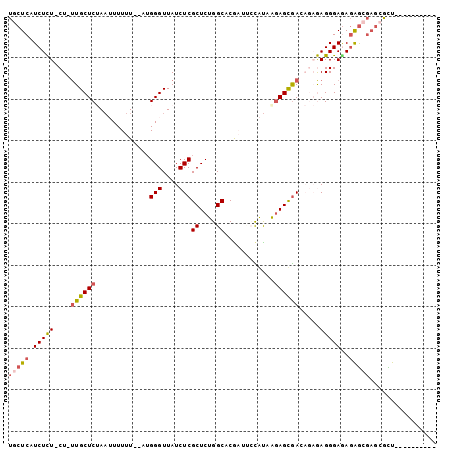
>2R_DroMel_CAF1 17749726 91 + 20766785 UGCUCAUCUCU-CU-UUGCUCUAAUUUUUUCGAUGGGUUAUCUCGCUCUGGCACGAGUCCAUUGGAGUGACAGAGAGGGAGAGAGAGAGCGCU---------- .((((.(((((-((-((.((((.....((((((((((....((((........))))))))))))))......)))))))))))))))))...---------- ( -37.20) >DroVir_CAF1 25374 97 + 1 UGCUCAUCUUUGCUUUUGCUCAAAAUUUUU--AUGGGUUAUCUCGCUCUGGCACGUUCGUGCAAGAGCAA----AAGGGAAAGAGCGAGAGCUGCCCGACAAU ...........((((((((((.........--..(((....)))(((((.((((....)))).)))))..----........))))))))))........... ( -32.70) >DroGri_CAF1 24768 94 + 1 UGCUCAUCUUUGCU-UUGCUCAACAUUUUU--AUGGGUUAUCUCGCUCUGGCACGCAAGUGCAAGAGCGA----AAGGGAGAGAGCGAGCACUGCCCGA--AU (((((.(((((.((-(((((((........--.)))))....(((((((.((((....)))).)))))))----)))).)))))..)))))........--.. ( -38.80) >DroSec_CAF1 17555 87 + 1 UGCUCAUCUCU-CU-UUGCUCUAAUUUUUU----GGGUUAUCUCGCUCUGGCACGAUUCCAUUGGAGUGACAGAGAGGGAGAGAAAGAGCGUU---------- .((((.(((((-((-((.((((.(((((.(----(((..(((..((....))..)))))))..)))))...)))))))))))))..))))...---------- ( -28.80) >DroEre_CAF1 18505 91 + 1 UGCUCAUCUCU-CU-UUGCUCUAAUUUUUUCGAUGGGUUAUCUCGCUCUGGCACGAUUCCAUAGCAGUGACAGAGAGGGAGAGAGCGAGCGCU---------- .((((.(((((-((-((.((((............(((....)))(((.(((.......))).)))......)))))))))))))..))))...---------- ( -30.90) >DroAna_CAF1 17713 87 + 1 UGCUCAUCUCU-CU-GUGCUCUAAUUUUUUCCAUGGGUUAUCUCGCUCUGGCACAACACCAUAAGAGCGAGAGAGAGGGAAAUGGC----GUU---------- .(((..(((((-((-..((((.............))))..(((((((((((.......))...))))))))).)))))))...)))----...---------- ( -27.92) >consensus UGCUCAUCUCU_CU_UUGCUCUAAUUUUUU__AUGGGUUAUCUCGCUCUGGCACGAUUCCAUAAGAGCGACAGAGAGGGAGAGAGCGAGCGCU__________ (((((.(((((....((((((.............(((....)))((....))............)))))).....)))))..)))))................ (-16.28 = -16.70 + 0.42)

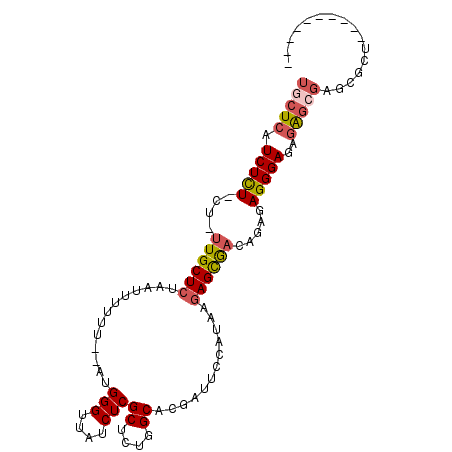
| Location | 17,749,726 – 17,749,817 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 74.71 |
| Mean single sequence MFE | -28.11 |
| Consensus MFE | -6.53 |
| Energy contribution | -6.03 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.23 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.721768 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17749726 91 - 20766785 ----------AGCGCUCUCUCUCUCCCUCUCUGUCACUCCAAUGGACUCGUGCCAGAGCGAGAUAACCCAUCGAAAAAAUUAGAGCAA-AG-AGAGAUGAGCA ----------...(((((((((((....(((((........((((.(((((......))))).....)))).........)))))...-))-))))).)))). ( -28.93) >DroVir_CAF1 25374 97 - 1 AUUGUCGGGCAGCUCUCGCUCUUUCCCUU----UUGCUCUUGCACGAACGUGCCAGAGCGAGAUAACCCAU--AAAAAUUUUGAGCAAAAGCAAAGAUGAGCA .(..(((((.(((....))).......((----(((((((.((((....)))).)))))))))...)))..--...........((....))...))..)... ( -27.90) >DroGri_CAF1 24768 94 - 1 AU--UCGGGCAGUGCUCGCUCUCUCCCUU----UCGCUCUUGCACUUGCGUGCCAGAGCGAGAUAACCCAU--AAAAAUGUUGAGCAA-AGCAAAGAUGAGCA ..--..((((...))))(((((((...((----(((((((.((((....)))).)))))))))........--.....((((......-)))).))).)))). ( -32.40) >DroSec_CAF1 17555 87 - 1 ----------AACGCUCUUUCUCUCCCUCUCUGUCACUCCAAUGGAAUCGUGCCAGAGCGAGAUAACCC----AAAAAAUUAGAGCAA-AG-AGAGAUGAGCA ----------...((((..((((((.(.(((((.((((((...)))...))).))))).).(......)----...............-.)-))))).)))). ( -24.00) >DroEre_CAF1 18505 91 - 1 ----------AGCGCUCGCUCUCUCCCUCUCUGUCACUGCUAUGGAAUCGUGCCAGAGCGAGAUAACCCAUCGAAAAAAUUAGAGCAA-AG-AGAGAUGAGCA ----------...(((((((((((....(((((......(.((((.(((.(((....))).)))...)))).).......)))))...-))-)))).))))). ( -27.72) >DroAna_CAF1 17713 87 - 1 ----------AAC----GCCAUUUCCCUCUCUCUCGCUCUUAUGGUGUUGUGCCAGAGCGAGAUAACCCAUGGAAAAAAUUAGAGCAC-AG-AGAGAUGAGCA ----------...----((((((((.((((((((((((((...(((.....))))))))))))...((...)).........)))...-))-.)))))).)). ( -27.70) >consensus __________AGCGCUCGCUCUCUCCCUCUCUGUCACUCCUAUGGAAUCGUGCCAGAGCGAGAUAACCCAU__AAAAAAUUAGAGCAA_AG_AGAGAUGAGCA .....................((((.((((....(((............)))..)))).))))........................................ ( -6.53 = -6.03 + -0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:48:38 2006