
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,691,738 – 17,691,858 |
| Length | 120 |
| Max. P | 0.999848 |

| Location | 17,691,738 – 17,691,858 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.50 |
| Mean single sequence MFE | -47.00 |
| Consensus MFE | -38.46 |
| Energy contribution | -38.38 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.982421 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
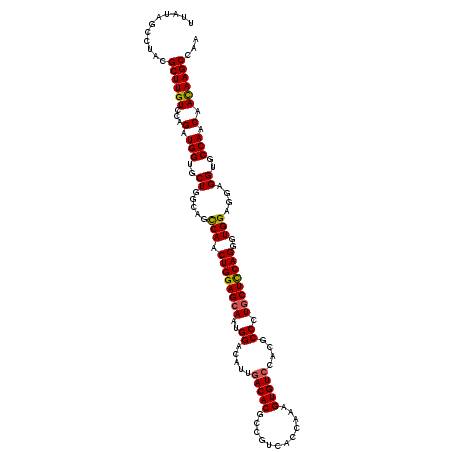
>2R_DroMel_CAF1 17691738 120 + 20766785 UUAUAGCCUACGCUUGUCCAGAUGGUGCUGGCAGCCAACUGGAGCAAUGGACAUUGACACGCCGUCACCAAAGUGUCCACGCCCUUCUCCAGGGUGGAGGAGGUGCCAACAACAAGCCAA ...........((((((((((.((((.......)))).)))))....(((((((((((.....)))).....))))))).((((((((((.....)))))))).))......)))))... ( -45.40) >DroSec_CAF1 1360 120 + 1 UUAUAGCCUACGCUUGUCCAGAUGGUGCUGGCAGCCAACUGGAGCAAUGGACAUUGACACGCCGUCACCAAAGUGUCCACACCCUGCUCCAGGGUGGCGGAGGAGCCAACAAUAAGCCAA ...........((((((...(.((((.((....((((.((((((((.(((((((((((.....)))).....))))))).....))))))))..))))...)).)))).).))))))... ( -43.90) >DroSim_CAF1 557 120 + 1 UUAUAGCCUACGCUUGUCCAGAUGGUGCUGGCAGCCAACUGGAGCAAUGGACAUUGACACGCCGUCACCAAAGUGUCCACGCCCUGCUUCAGGGUGGCGGAGGUGCCAACAACAAGCCAA ...........((((((...(.(((..((....((((.((((((((.(((((((((((.....)))).....))))))).....))))))))..))))...))..))).).))))))... ( -46.40) >DroEre_CAF1 1178 120 + 1 UCAUAGCCUACGCUUGUCCGGAUGGUGCUGGAAGCCAACUGGAGCAAUGGACGUUGACACGCCGGCACCAAAGUGUCCACGCCCUGCUCCAGGGUGGAGGAGGUGCCAACAACAAGCCAA ...........((((((...(.(((..((.....(((.((((((((..((.(((.(((((...(....)...))))).))))).))))))))..)))....))..))).).))))))... ( -49.80) >DroYak_CAF1 1057 120 + 1 UCAUAGCCUACGCUUGUCCGGAUGGUGCUGGCAGUCAACUGGAGCAGUGGACGUUGACACGCCGGCACCAAAGUGUGCACGCCCUGCUCCAGGGUGGAGGAGGUGCCAACAACAAGCCAA ...........((((((...(.(((..((..(..(((.(((((((((.((.(((.(.(((((.(....)...))))))))))))))))))))..)))..).))..))).).))))))... ( -49.50) >consensus UUAUAGCCUACGCUUGUCCAGAUGGUGCUGGCAGCCAACUGGAGCAAUGGACAUUGACACGCCGUCACCAAAGUGUCCACGCCCUGCUCCAGGGUGGAGGAGGUGCCAACAACAAGCCAA ...........((((((...(.(((..((.....(((.((((((((..((.....(((((............)))))....)).))))))))..)))....))..))).).))))))... (-38.46 = -38.38 + -0.08)

| Location | 17,691,738 – 17,691,858 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.50 |
| Mean single sequence MFE | -50.82 |
| Consensus MFE | -46.30 |
| Energy contribution | -47.30 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.91 |
| SVM decision value | 4.24 |
| SVM RNA-class probability | 0.999848 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17691738 120 - 20766785 UUGGCUUGUUGUUGGCACCUCCUCCACCCUGGAGAAGGGCGUGGACACUUUGGUGACGGCGUGUCAAUGUCCAUUGCUCCAGUUGGCUGCCAGCACCAUCUGGACAAGCGUAGGCUAUAA ...((((((((((((((......(((..((((((..((((((.(((((.(((....))).))))).))))))....)))))).))).))))))))((....))))))))........... ( -50.80) >DroSec_CAF1 1360 120 - 1 UUGGCUUAUUGUUGGCUCCUCCGCCACCCUGGAGCAGGGUGUGGACACUUUGGUGACGGCGUGUCAAUGUCCAUUGCUCCAGUUGGCUGCCAGCACCAUCUGGACAAGCGUAGGCUAUAA .(((((((((((((((......((((..(((((((((((..(.(((((.(((....))).))))).)..)))..)))))))).)))).)))))))((....))......))))))))... ( -54.10) >DroSim_CAF1 557 120 - 1 UUGGCUUGUUGUUGGCACCUCCGCCACCCUGAAGCAGGGCGUGGACACUUUGGUGACGGCGUGUCAAUGUCCAUUGCUCCAGUUGGCUGCCAGCACCAUCUGGACAAGCGUAGGCUAUAA ...((((((((((((((.....((((..(((.((((((((((.(((((.(((....))).))))).))))))..)))).))).))))))))))))((....))))))))........... ( -52.40) >DroEre_CAF1 1178 120 - 1 UUGGCUUGUUGUUGGCACCUCCUCCACCCUGGAGCAGGGCGUGGACACUUUGGUGCCGGCGUGUCAACGUCCAUUGCUCCAGUUGGCUUCCAGCACCAUCCGGACAAGCGUAGGCUAUGA ...((((((((((((........(((..((((((((((((((.(((((.(((....))).))))).))))))..)))))))).)))...))))))((....))))))))........... ( -48.70) >DroYak_CAF1 1057 120 - 1 UUGGCUUGUUGUUGGCACCUCCUCCACCCUGGAGCAGGGCGUGCACACUUUGGUGCCGGCGUGUCAACGUCCACUGCUCCAGUUGACUGCCAGCACCAUCCGGACAAGCGUAGGCUAUGA ...(((((((..(((((((((((((.....))))..))).)))).))...((((((.((((.((((((.............)))))))))).))))))....)))))))........... ( -48.12) >consensus UUGGCUUGUUGUUGGCACCUCCUCCACCCUGGAGCAGGGCGUGGACACUUUGGUGACGGCGUGUCAAUGUCCAUUGCUCCAGUUGGCUGCCAGCACCAUCUGGACAAGCGUAGGCUAUAA ...((((((((((((((......(((..((((((((((((((.(((((.(((....))).))))).))))))..)))))))).))).))))))))((....))))))))........... (-46.30 = -47.30 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:48:20 2006