
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,686,770 – 17,686,860 |
| Length | 90 |
| Max. P | 0.855567 |

| Location | 17,686,770 – 17,686,860 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 80.16 |
| Mean single sequence MFE | -36.60 |
| Consensus MFE | -20.23 |
| Energy contribution | -24.11 |
| Covariance contribution | 3.88 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.637712 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
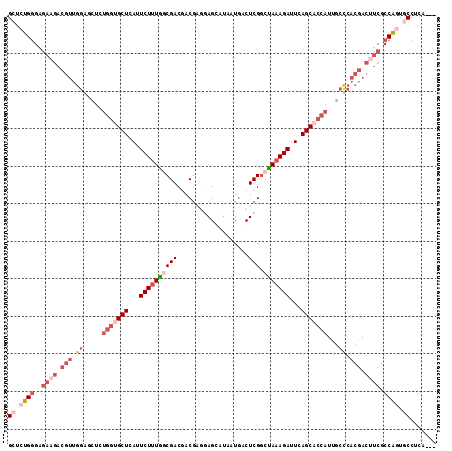
>2R_DroMel_CAF1 17686770 90 + 20766785 GCUCUGUG---------------------GCUCAUUCUUUGGCGACGACGAGGAGCAUAAUGACUCGGCUAAAGAUUCAGCACCAUUGCCCACGACUUCGCCAGUGCCUCA--- .....(.(---------------------((.......(((((((.(.((.((.(((...(((.((.......)).))).......))))).)).).))))))).))).).--- ( -24.10) >DroSec_CAF1 3844 111 + 1 GCUCUGGGAGAAGACGUUGGGGCUAUGGCGCUCAUUCUUUAACGACGACAAGGAGGAUAAUGACUCGGCUAUAGAUUCAGCACCAUUGCCCACGACUUCGCCAGUGCCUCA--- ((.((((..((((.(((..((((.((((.(((...(((.((.(..(.....)(((........))).).)).)))...))).)))).))))))).)))).)))).))....--- ( -37.40) >DroSim_CAF1 871 114 + 1 GCUCUGGGAGAAGACGUUGGGGCUGUGGUGCUCAUUCUUUAACGACGACGAGGAGGAUAAUGACUCGGCUAAAGAUUCAGCACCAUUGCCCACGACUUCGCCAGUGCCUCAGCC ((.((((..((((.(((..((((.((((((((...((((((..(....((((..(.....)..)))).)))))))...)))))))).))))))).)))).)))).))....... ( -46.20) >DroEre_CAF1 4604 108 + 1 GCUCUGGGAGAAGACGUUGGAUUUCUGGUGCUCAUUCUUUGGCGACCACG---AGCAUAAUGGCUCGGCCAAAGAUUCAGCACCAUCACUCACGACGUCGCCGAUACCUCA--- ......((....(((((((((....(((((((...((((((((.....((---(((......)))))))))))))...)))))))....)).))))))).)).........--- ( -43.90) >DroYak_CAF1 3810 108 + 1 GCUCUGGGAGAAGACGUUGGAUUUCUGGUGCUCAUUCUUUGGCGACCACG---AGCAUAAUUACUCGACUAAAGAUUCAGCACCAUCGCUCACGACUUCGCCAAUACCUCA--- ......((.((((.(((.((.....(((((((...(((((((......((---((........)))).)))))))...)))))))...)).))).)))).)).........--- ( -31.40) >consensus GCUCUGGGAGAAGACGUUGGAGCUCUGGUGCUCAUUCUUUGGCGACGACGAGGAGCAUAAUGACUCGGCUAAAGAUUCAGCACCAUUGCCCACGACUUCGCCAGUGCCUCA___ ((.((((..((((.(((.((.....(((((((...((((((((((...................))).)))))))...)))))))...)).))).)))).)))).))....... (-20.23 = -24.11 + 3.88)

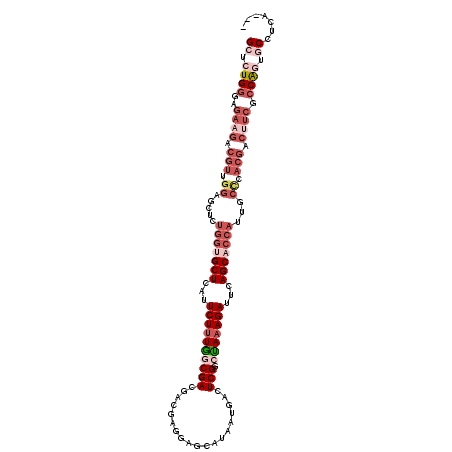
| Location | 17,686,770 – 17,686,860 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 80.16 |
| Mean single sequence MFE | -38.82 |
| Consensus MFE | -23.31 |
| Energy contribution | -25.79 |
| Covariance contribution | 2.48 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.855567 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
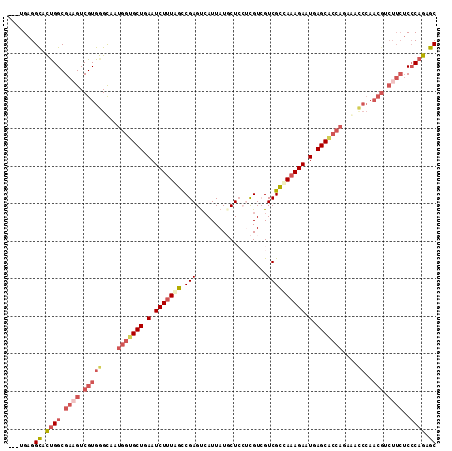
>2R_DroMel_CAF1 17686770 90 - 20766785 ---UGAGGCACUGGCGAAGUCGUGGGCAAUGGUGCUGAAUCUUUAGCCGAGUCAUUAUGCUCCUCGUCGUCGCCAAAGAAUGAGC---------------------CACAGAGC ---((.(((.(((((((.(.((.(((((((((.(((((....)))))....))))..))).)).)).).))))))......).))---------------------).)).... ( -27.00) >DroSec_CAF1 3844 111 - 1 ---UGAGGCACUGGCGAAGUCGUGGGCAAUGGUGCUGAAUCUAUAGCCGAGUCAUUAUCCUCCUUGUCGUCGUUAAAGAAUGAGCGCCAUAGCCCCAACGUCUUCUCCCAGAGC ---....((.((((.((((.(((((((.((((((((.(.(((.((((.(((.((..........)).).)))))).))).).)))))))).))))..))).))))..)))).)) ( -43.40) >DroSim_CAF1 871 114 - 1 GGCUGAGGCACUGGCGAAGUCGUGGGCAAUGGUGCUGAAUCUUUAGCCGAGUCAUUAUCCUCCUCGUCGUCGUUAAAGAAUGAGCACCACAGCCCCAACGUCUUCUCCCAGAGC .......((.((((.((((.(((((((..(((((((.(.((((((((((((...........)))).....)))))))).).)))))))..))))..))).))))..)))).)) ( -46.10) >DroEre_CAF1 4604 108 - 1 ---UGAGGUAUCGGCGACGUCGUGAGUGAUGGUGCUGAAUCUUUGGCCGAGCCAUUAUGCU---CGUGGUCGCCAAAGAAUGAGCACCAGAAAUCCAACGUCUUCUCCCAGAGC ---((.((....((.(((((.(.((....(((((((.(.(((((((((((((......)))---)).....)))))))).).)))))))....))).))))).)).)))).... ( -42.20) >DroYak_CAF1 3810 108 - 1 ---UGAGGUAUUGGCGAAGUCGUGAGCGAUGGUGCUGAAUCUUUAGUCGAGUAAUUAUGCU---CGUGGUCGCCAAAGAAUGAGCACCAGAAAUCCAACGUCUUCUCCCAGAGC ---....((.((((.((((.(((((....(((((((.(.(((((.((((((((....))))---)).....)).))))).).)))))))....))..))).))))..)))).)) ( -35.40) >consensus ___UGAGGCACUGGCGAAGUCGUGGGCAAUGGUGCUGAAUCUUUAGCCGAGUCAUUAUGCUCCUCGUCGUCGCCAAAGAAUGAGCACCAGAAACCCAACGUCUUCUCCCAGAGC .......((.((((.((((.(((((....(((((((.(.(((((((.(((...................)))))))))).).)))))))....))..))).))))..)))).)) (-23.31 = -25.79 + 2.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:48:14 2006