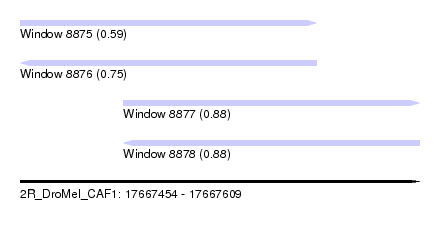
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,667,454 – 17,667,609 |
| Length | 155 |
| Max. P | 0.884586 |

| Location | 17,667,454 – 17,667,569 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 89.80 |
| Mean single sequence MFE | -40.77 |
| Consensus MFE | -29.46 |
| Energy contribution | -29.79 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.592387 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17667454 115 + 20766785 AUGCGAUGCGGCAUCAGCGUCGGCAACAUUCCAGCCAUGAGUCACUGCAGCAGAAGCAACAGCAGCAGC----ACACUUGCUGCGCCGCUGCUGCGACAUGGGGAUAAUCACAUUGAUA .((((((((.......))))).)))....(((..(((((......((((((((..((....)).(((((----(....))))))....)))))))).))))))))..(((.....))). ( -40.30) >DroSec_CAF1 20852 119 + 1 AUGCGAUGCGGCAUCAGCGUCGGCAACAUUCCAGCCAUGAGUCACUGCAGCAGAAGCAACAUCAGCAGCAGCAACACUUGCUGCGCCGCUUCUGCGACAUGGGGAUAAUCACAUUGAUA .((((((((.......))))).)))....(((..(((((.((....)).((((((((.......(((((((......)))))))...))))))))..))))))))..(((.....))). ( -40.80) >DroEre_CAF1 21303 107 + 1 AUGCGAUGCGGCAUCAGCUUCGGCAACAUUCCAGCCAUGAGUCACAGCAGCAGA---AGCAGCA---------ACACUUGCUGCACCGCUGCUGCGACAUGGGGAUAAUCACAUUGAUA ...(((((.((.....((....)).....(((..((((..(((.(((((((.(.---.((((((---------.....))))))..)))))))).))))))))))...)).)))))... ( -41.20) >consensus AUGCGAUGCGGCAUCAGCGUCGGCAACAUUCCAGCCAUGAGUCACUGCAGCAGAAGCAACAGCAGCAGC____ACACUUGCUGCGCCGCUGCUGCGACAUGGGGAUAAUCACAUUGAUA .((((((((.......))))).)))....(((..(((((.......(((((((........................)))))))..(((....))).))))))))..(((.....))). (-29.46 = -29.79 + 0.33)


| Location | 17,667,454 – 17,667,569 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 89.80 |
| Mean single sequence MFE | -45.93 |
| Consensus MFE | -34.09 |
| Energy contribution | -34.54 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.751237 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17667454 115 - 20766785 UAUCAAUGUGAUUAUCCCCAUGUCGCAGCAGCGGCGCAGCAAGUGU----GCUGCUGCUGUUGCUUCUGCUGCAGUGACUCAUGGCUGGAAUGUUGCCGACGCUGAUGCCGCAUCGCAU .....((((((...((((((((((((((((((((((((.....)))----)))))))))((.((....)).)).)))))..))))..)))....(((....((....)).))))))))) ( -43.90) >DroSec_CAF1 20852 119 - 1 UAUCAAUGUGAUUAUCCCCAUGUCGCAGAAGCGGCGCAGCAAGUGUUGCUGCUGCUGAUGUUGCUUCUGCUGCAGUGACUCAUGGCUGGAAUGUUGCCGACGCUGAUGCCGCAUCGCAU ......((((((.........))))))((.((((((((((..(.((..((((.((.((.......)).)).))))..)).).((((.........))))..)))).)))))).)).... ( -46.00) >DroEre_CAF1 21303 107 - 1 UAUCAAUGUGAUUAUCCCCAUGUCGCAGCAGCGGUGCAGCAAGUGU---------UGCUGCU---UCUGCUGCUGUGACUCAUGGCUGGAAUGUUGCCGAAGCUGAUGCCGCAUCGCAU .....((((((...((((((((((((((((((((.((((((.....---------)))))).---.)))))))))))))..))))..)))....(((....((....)).))))))))) ( -47.90) >consensus UAUCAAUGUGAUUAUCCCCAUGUCGCAGCAGCGGCGCAGCAAGUGU____GCUGCUGCUGUUGCUUCUGCUGCAGUGACUCAUGGCUGGAAUGUUGCCGACGCUGAUGCCGCAUCGCAU .....((((((...((((((((((((.(((((((.((((((..............)))))).....))))))).)))))..))))..)))....(((....((....)).))))))))) (-34.09 = -34.54 + 0.45)

| Location | 17,667,494 – 17,667,609 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 89.24 |
| Mean single sequence MFE | -41.87 |
| Consensus MFE | -30.50 |
| Energy contribution | -31.50 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.46 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878044 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17667494 115 + 20766785 GUCACUGCAGCAGAAGCAACAGCAGCAGC----ACACUUGCUGCGCCGCUGCUGCGACAUGGGGAUAAUCACAUUGAUAUGCCUAAUUAGCUGCAACAGCGACCACUGCCUAACUGGCC (((.(((..((((..(((.((((.(((((----(....))))))...)))).)))....((((.(((.((.....))))).)))).....))))..))).)))....(((.....))). ( -43.40) >DroSec_CAF1 20892 119 + 1 GUCACUGCAGCAGAAGCAACAUCAGCAGCAGCAACACUUGCUGCGCCGCUUCUGCGACAUGGGGAUAAUCACAUUGAUAUGCCUAAUUAGCUGCAACAGCGACCACUGCCUAACUGGCC (((..((..((((((((.......(((((((......)))))))...))))))))..))((((.(((.((.....))))).))))....((((...)))))))....(((.....))). ( -40.60) >DroEre_CAF1 21343 107 + 1 GUCACAGCAGCAGA---AGCAGCA---------ACACUUGCUGCACCGCUGCUGCGACAUGGGGAUAAUCACAUUGAUAUGCCUAAUUAGCUGCAACAGCGACUACUGCCUAACUGGCU (((.(((((((.(.---.((((((---------.....))))))..)))))))).))).((((.(((.((.....))))).))))....((((...)))).......(((.....))). ( -41.60) >consensus GUCACUGCAGCAGAAGCAACAGCAGCAGC____ACACUUGCUGCGCCGCUGCUGCGACAUGGGGAUAAUCACAUUGAUAUGCCUAAUUAGCUGCAACAGCGACCACUGCCUAACUGGCC (((...(((((((......((((((............)))))).....)))))))....((((.(((.((.....))))).))))....((((...)))))))....(((.....))). (-30.50 = -31.50 + 1.00)

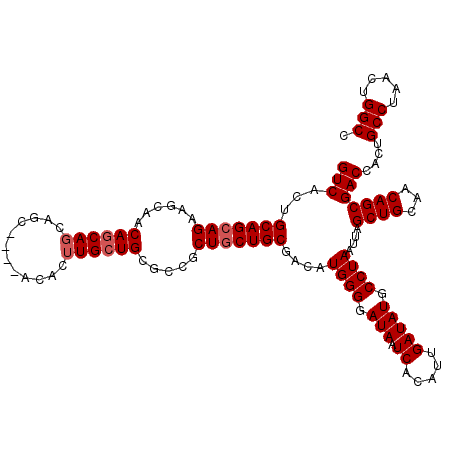
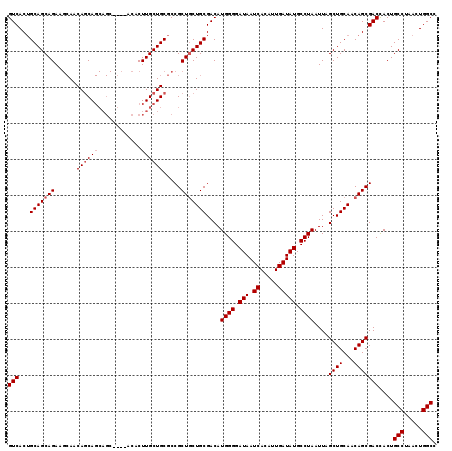
| Location | 17,667,494 – 17,667,609 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 89.24 |
| Mean single sequence MFE | -49.77 |
| Consensus MFE | -35.53 |
| Energy contribution | -37.98 |
| Covariance contribution | 2.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.88 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.884586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17667494 115 - 20766785 GGCCAGUUAGGCAGUGGUCGCUGUUGCAGCUAAUUAGGCAUAUCAAUGUGAUUAUCCCCAUGUCGCAGCAGCGGCGCAGCAAGUGU----GCUGCUGCUGUUGCUUCUGCUGCAGUGAC .(((.....)))....((((((((.((((.......((...(((.....)))...))....((.((((((((((((((.....)))----))))))))))).))..)))).)))))))) ( -51.10) >DroSec_CAF1 20892 119 - 1 GGCCAGUUAGGCAGUGGUCGCUGUUGCAGCUAAUUAGGCAUAUCAAUGUGAUUAUCCCCAUGUCGCAGAAGCGGCGCAGCAAGUGUUGCUGCUGCUGAUGUUGCUUCUGCUGCAGUGAC .(((.....)))....((((((((.((((......(((((((((..((((((.........))))))..(((((((((((....))))).)))))))))).))))))))).)))))))) ( -49.60) >DroEre_CAF1 21343 107 - 1 AGCCAGUUAGGCAGUAGUCGCUGUUGCAGCUAAUUAGGCAUAUCAAUGUGAUUAUCCCCAUGUCGCAGCAGCGGUGCAGCAAGUGU---------UGCUGCU---UCUGCUGCUGUGAC .(((.....))).((((((((.((((..(((.....)))....))))))))))))......(((((((((((((.((((((.....---------)))))).---.))))))))))))) ( -48.60) >consensus GGCCAGUUAGGCAGUGGUCGCUGUUGCAGCUAAUUAGGCAUAUCAAUGUGAUUAUCCCCAUGUCGCAGCAGCGGCGCAGCAAGUGU____GCUGCUGCUGUUGCUUCUGCUGCAGUGAC .(((.....)))....((((((((.((((.......((((((((.....))).......)))))(((((((((((...((....))....))))))))))).....)))).)))))))) (-35.53 = -37.98 + 2.45)

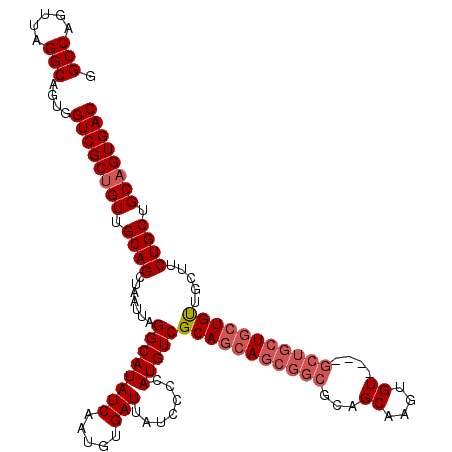
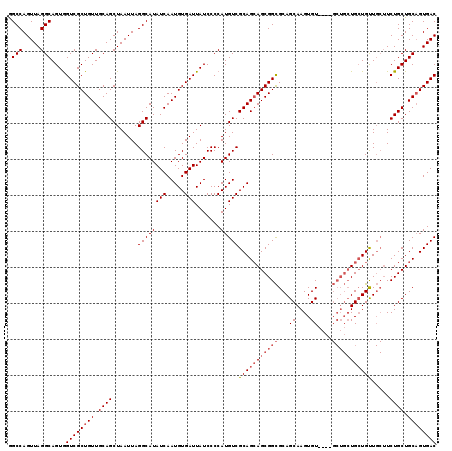
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:48:06 2006