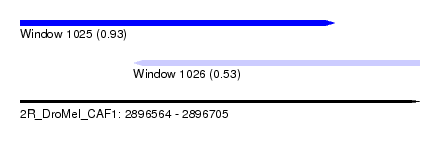
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,896,564 – 2,896,705 |
| Length | 141 |
| Max. P | 0.929779 |

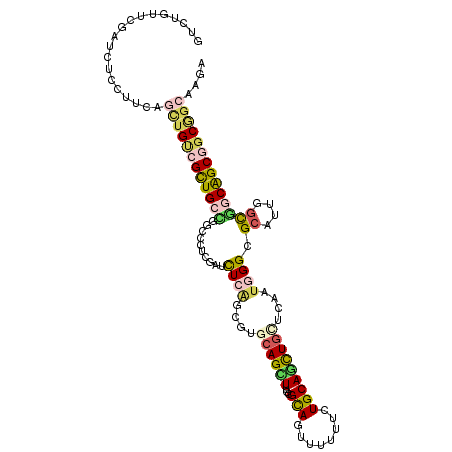
| Location | 2,896,564 – 2,896,675 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 77.12 |
| Mean single sequence MFE | -42.80 |
| Consensus MFE | -24.50 |
| Energy contribution | -25.28 |
| Covariance contribution | 0.79 |
| Combinations/Pair | 1.56 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2896564 111 + 20766785 GUCUCUUCGAUUUCCCGUAACUGUCGUUGCUUGCCCUCGAUCUCAGCGACCAGUUUGCGCAAUUUUUUCUGCAGCUGUUCAAUAGGAGCAGUGGCGGCGGCGGCAGCGAGA .((((..((......))...(((((((((((.(((..........((((.....))))(((........))).(((((((.....))))))))))))))))))))).)))) ( -37.70) >DroVir_CAF1 1544 108 + 1 GUCUGUUCGAUUUCAUUCAGCUGUCGCUGACGCGCCUCGAUUUCGGCGUGCAGCUUUCGCACCUUUUUCUGCACCUGCUCAAUGGGCGCAUUUGCGCCAGCAGCGCCG--- (.(((((......((((.(((....((((.((((((........))))))))))....(((........)))....))).))))(((((....)))))))))))....--- ( -39.80) >DroPse_CAF1 1601 111 + 1 GUCUGCUCGAUCUCCUUCAGCUGCCGCUGCCGUCCCUCGAUCUCAGCGUGCAGCUUUCGCAGUUUCUUCUGCAGCUGCUCGAUGGGCGCAUUGGCGGCAGCGGCGGCAAUG ........((......)).(((((((((((((((...((..((((.((.((((((...((((......)))))))))).)).))))))....))))))))))))))).... ( -56.00) >DroEre_CAF1 1556 111 + 1 GUCUCUUCGAUUUCCCGCAACUGUCGUUGCUUGCCCUCGAUCUCAGCGACCAGUUUGCGCAAUUUUUUCUGCAACUGCUCAAUAGGCGCAGUGGCAGCGGCGGCAGCGAGA .((((..((......))...(((((((((((.(((((((.(((....((.(((((.(((.((....)).)))))))).))...))))).)).)))))))))))))).)))) ( -36.80) >DroWil_CAF1 1520 111 + 1 GUCUGUUCGAUCUCCUUAAGUUGACGCUGGCGACCCUCAAUCUCUGCAUGAAGUUUGCGUAGUUUUUUCUGCAGUUGUUCAAUGGGAGCAUUGAUAGCGGCAGCUGCAGCU ..................(((((..((((.((.(..((((((((((((.......)))((((......))))............)))).)))))..))).))))..))))) ( -30.50) >DroPer_CAF1 1601 111 + 1 GUCUGCUCGAUCUCCUUCAGCUGCCGCUGCCGUCCCUCGAUCUCAGCGUGCAGCUUUCGCAGUUUCUUCUGCAGCUGCUCGAUGGGCGCAUUGGCGGCAGCGGCGGCAAUG ........((......)).(((((((((((((((...((..((((.((.((((((...((((......)))))))))).)).))))))....))))))))))))))).... ( -56.00) >consensus GUCUGUUCGAUCUCCUUCAGCUGUCGCUGCCGGCCCUCGAUCUCAGCGUGCAGCUUGCGCAGUUUUUUCUGCAGCUGCUCAAUGGGCGCAUUGGCGGCAGCGGCGGCAAGA ...................((((((((((((..........((((....((((((...(((........)))))))))....)))).((....)))))))))))))).... (-24.50 = -25.28 + 0.79)

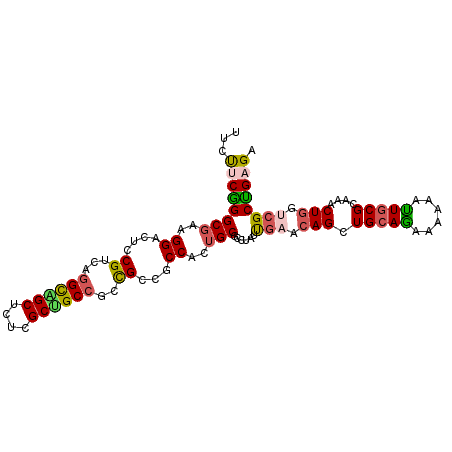
| Location | 2,896,604 – 2,896,705 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 81.19 |
| Mean single sequence MFE | -40.32 |
| Consensus MFE | -23.11 |
| Energy contribution | -22.83 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.34 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.57 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.525422 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2896604 101 - 20766785 UUCUUCGGGCGAAGGAAUCCGUCAGGCGGCUCUCGCUGCCGCCGCCGCCACUGCUCCUAUUGAACAGCUGCAGAAAAAAUUGCGCAAACUGGUCGCUGAGA ((((((....)))))).....(((((((((.......))))))((.(((((((.((.....)).)))..((((......))))......)))).))))).. ( -30.50) >DroPse_CAF1 1641 101 - 1 CUCCGCAGGCGAAGGGCUGCGGCAGGCGGCCAUUGCCGCCGCUGCCGCCAAUGCGCCCAUCGAGCAGCUGCAGAAGAAACUGCGAAAGCUGCACGCUGAGA (((.((.((((...(((.(((((.((((((....))))))))))).)))....))))......((((((((((......))))...))))))..)).))). ( -56.40) >DroSec_CAF1 1461 101 - 1 UUCUUCGGGCGAAGGACUUCGUCAAGCAGCUCUCGCAGCGGCCGCUGCCACAGCUCCAACUGAACAGCUGCAGAAAAAAUUACGCAAACUGGUCGCCGAGA .......((((((....)))))).......(((((..((((((((((...(((......)))..))))(((............)))....))))))))))) ( -30.70) >DroEre_CAF1 1596 101 - 1 UUCUUCGGGCGAGGGACUCCGUCAGGCAGCUCUCGCUGCCGCCGCUGCCACUGCGCCUAUUGAGCAGUUGCAGAAAAAAUUGCGCAAACUGGUCGCUGAGA .((((..(((((.((...)).)).((((((....))))))))).((((.(((((.........))))).))))........((((......).))).)))) ( -37.10) >DroYak_CAF1 1575 101 - 1 UUCUUCGGGCGAGGGACUUCGUCAGGCAGCUCUCGCUGCCGCCGCUGCCACUGCUCCUAUUGAACAGCUGCAAAAAAAAUUGCGCAAACUAGUAGCCGAGA ...(((((((((.(.....).)).((((((....)))))))))(((((..(((.((.....)).)))..((((......))))........))))))))). ( -32.70) >DroPer_CAF1 1641 101 - 1 CUCCGCAGGCGAAGGGCUGCGGCAGGUGGCCAUUGCCGCCGCUGCCGCCAAUGCGCCCAUCGAGCAGCUGCAGAAGAAACUGCGAAAGCUGCACGCUGAGA (((.((.((((...(((.(((((.((((((....))))))))))).)))....))))......((((((((((......))))...))))))..)).))). ( -54.50) >consensus UUCUUCGGGCGAAGGACUCCGUCAGGCAGCUCUCGCUGCCGCCGCCGCCACUGCGCCUAUUGAACAGCUGCAGAAAAAAUUGCGCAAACUGGUCGCUGAGA ...((((((((..((....((...((((((....))))))..))...))..)))......(((.(((.(((((......)))))....))).)))))))). (-23.11 = -22.83 + -0.27)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:41:34 2006