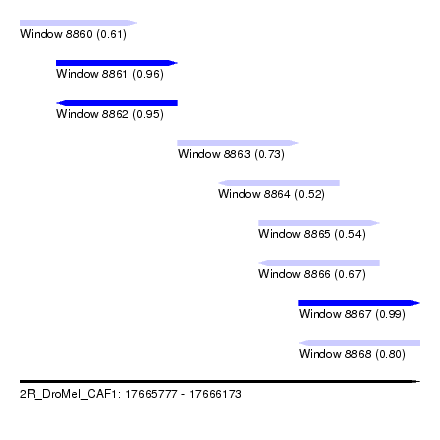
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,665,777 – 17,666,173 |
| Length | 396 |
| Max. P | 0.986198 |

| Location | 17,665,777 – 17,665,893 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.72 |
| Mean single sequence MFE | -34.00 |
| Consensus MFE | -21.31 |
| Energy contribution | -23.62 |
| Covariance contribution | 2.31 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.612508 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17665777 116 + 20766785 CCUUUCACUUUUCAACCUGGG----UACUCCUUGGACCUGACACGUUGGAAGCAGAGUGCCAUCGUCUAGAUCACAGCUGCAACAUGCAGCACAUUCCCCGAUUGCACAUGUGCCCUCAU ..................(((----(((...((((((.((.(((.(((....))).))).))..))))))......(((((.....)))))...................)))))).... ( -30.10) >DroSec_CAF1 19212 116 + 1 CCUUUCACUUUUCAACCUGGG----UACUCCUUGGACCUGACACCUUGGAAGCAGAGUGGCAUCGUCCAGAGCACAGCUGCAACAUGCAGCACAUUCCCCGAGUGCACAUGUGCCCUCAU ..................(((----(((...((((((.((.(((.(((....))).))).))..)))))).((((.(((((.....))))).(.......).))))....)))))).... ( -36.50) >DroEre_CAF1 19596 106 + 1 CCUUUCACUUUUCAACCUGGAGCCUAACUCCUAGGA--------------AGCAGAGUGGCAUCGUCCAAGGCGCAGCUGCAGCAUGCAGCACAUUCCCGGAGUGCACAUGUGCCCUCAU ....(((((((.(..(((((((.....)))).))).--------------.).)))))))..........(((((((((((.....))))).(((((...)))))....))))))..... ( -35.90) >DroYak_CAF1 31283 115 + 1 CCUUUCACUUUUCAACCUGGG----UACUCCUUGGACCCGACACCUUG-UGGCAGUGUAGCAUCGUCCAAAGCACAGCUGCAACAUGCAGCACAUUCCCGGAGUGCACAUGUGCCCUCAU ..................(((----(((...((((((....(((...)-))((......))...)))))).((((.(((((.....)))))...((....))))))....)))))).... ( -33.50) >consensus CCUUUCACUUUUCAACCUGGG____UACUCCUUGGACCUGACACCUUG_AAGCAGAGUGGCAUCGUCCAAAGCACAGCUGCAACAUGCAGCACAUUCCCCGAGUGCACAUGUGCCCUCAU ..................(((..........((((((....(((.(((....))).))).....))))))......(((((.....))))).....))).(((.(((....))).))).. (-21.31 = -23.62 + 2.31)

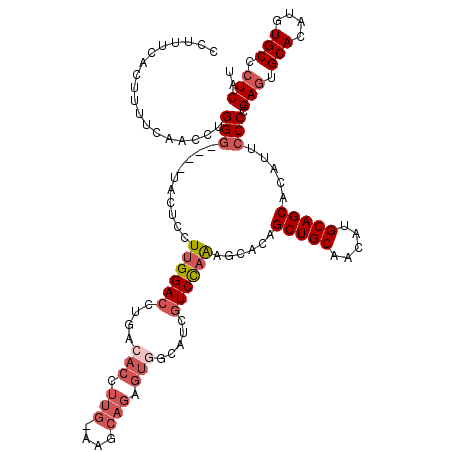
| Location | 17,665,813 – 17,665,933 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.43 |
| Mean single sequence MFE | -39.75 |
| Consensus MFE | -30.61 |
| Energy contribution | -33.17 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961513 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17665813 120 + 20766785 ACACGUUGGAAGCAGAGUGCCAUCGUCUAGAUCACAGCUGCAACAUGCAGCACAUUCCCCGAUUGCACAUGUGCCCUCAUAAUUGAGGUUCAGCCGCAUUCUGAGCACUCCUUUUCCAUU ......((((((..((((((((..............(((((.....))))).........((.(((...((...(((((....)))))..))...))).)))).))))))..)))))).. ( -39.00) >DroSec_CAF1 19248 120 + 1 ACACCUUGGAAGCAGAGUGGCAUCGUCCAGAGCACAGCUGCAACAUGCAGCACAUUCCCCGAGUGCACAUGUGCCCUCAUAAUUGAGGUUCAGCCGCAUUCUGAGCACUCCUUUUCCACU ......((((((..(((((((..........((((((((((.....))))).(((((...)))))....)))))(((((....)))))....)))((.......))))))..)))))).. ( -41.10) >DroEre_CAF1 19632 110 + 1 ----------AGCAGAGUGGCAUCGUCCAAGGCGCAGCUGCAGCAUGCAGCACAUUCCCGGAGUGCACAUGUGCCCUCAUAAUUGAGGUUCAGCCGCAUUCUGAGCACUCCUUUUCCACU ----------....(((((((..(......)..)).(((((.....))))).)))))..(((((((.((.(((((((((....))))).......))))..)).)))))))......... ( -39.51) >DroYak_CAF1 31319 119 + 1 ACACCUUG-UGGCAGUGUAGCAUCGUCCAAAGCACAGCUGCAACAUGCAGCACAUUCCCGGAGUGCACAUGUGCCCUCAUAAUUGAGGUUCAGCCGCAUUCUGAGCACUCCUUUUCCACU .......(-(((..((((.((((.((....(((...)))...)))))).))))......(((((((.((.(((((((((....))))).......))))..)).)))))))....)))). ( -39.41) >consensus ACACCUUG_AAGCAGAGUGGCAUCGUCCAAAGCACAGCUGCAACAUGCAGCACAUUCCCCGAGUGCACAUGUGCCCUCAUAAUUGAGGUUCAGCCGCAUUCUGAGCACUCCUUUUCCACU ......((((((..(((((.................(((((.....))))).......((((((((...((...(((((....)))))..))...))))))))..)))))..)))))).. (-30.61 = -33.17 + 2.56)

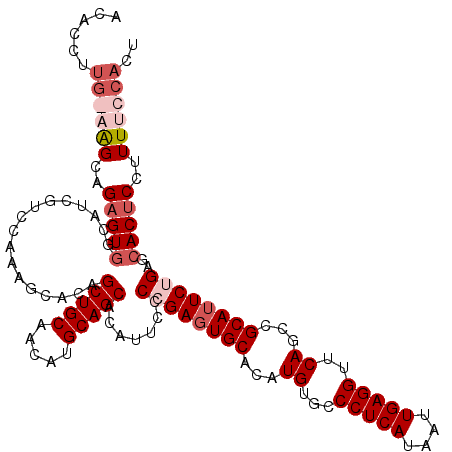
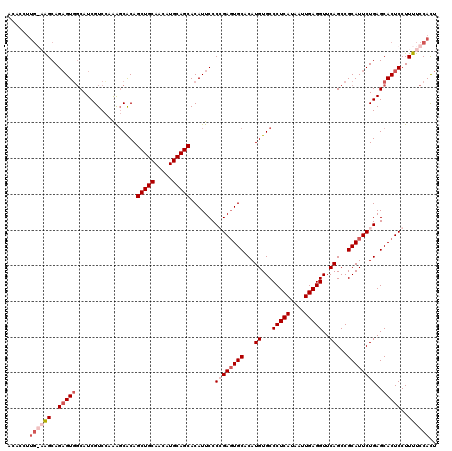
| Location | 17,665,813 – 17,665,933 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.43 |
| Mean single sequence MFE | -44.38 |
| Consensus MFE | -36.62 |
| Energy contribution | -38.75 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953135 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17665813 120 - 20766785 AAUGGAAAAGGAGUGCUCAGAAUGCGGCUGAACCUCAAUUAUGAGGGCACAUGUGCAAUCGGGGAAUGUGCUGCAUGUUGCAGCUGUGAUCUAGACGAUGGCACUCUGCUUCCAACGUGU ..(((((..((((((((((((.(((((((...(((((....)))))(((((((((((...........))).))))).)))))))))).))).)).....)))))))..)))))...... ( -44.70) >DroSec_CAF1 19248 120 - 1 AGUGGAAAAGGAGUGCUCAGAAUGCGGCUGAACCUCAAUUAUGAGGGCACAUGUGCACUCGGGGAAUGUGCUGCAUGUUGCAGCUGUGCUCUGGACGAUGCCACUCUGCUUCCAAGGUGU ..(((((..((((((((((((..((((((...(((((....)))))((((((((((((.........)))).))))).)))))))))..))))).....).))))))..)))))...... ( -48.70) >DroEre_CAF1 19632 110 - 1 AGUGGAAAAGGAGUGCUCAGAAUGCGGCUGAACCUCAAUUAUGAGGGCACAUGUGCACUCCGGGAAUGUGCUGCAUGCUGCAGCUGCGCCUUGGACGAUGCCACUCUGCU---------- ((..((...((..((..(((..(((((((...(((((....)))))((((((((((((.........)))).))))).))))))))))..)))..))...))..))..))---------- ( -41.50) >DroYak_CAF1 31319 119 - 1 AGUGGAAAAGGAGUGCUCAGAAUGCGGCUGAACCUCAAUUAUGAGGGCACAUGUGCACUCCGGGAAUGUGCUGCAUGUUGCAGCUGUGCUUUGGACGAUGCUACACUGCCA-CAAGGUGU .((((...((.((((..((((..((((((...(((((....)))))((((((((((((.........)))).))))).)))))))))..))))..)...)))...)).)))-)....... ( -42.60) >consensus AGUGGAAAAGGAGUGCUCAGAAUGCGGCUGAACCUCAAUUAUGAGGGCACAUGUGCACUCCGGGAAUGUGCUGCAUGUUGCAGCUGUGCUCUGGACGAUGCCACUCUGCUU_CAAGGUGU ..(((((..((((((((((((.(((((((...(((((....)))))((((((((((((.........)))).))))).)))))))))).))))).....).))))))..)))))...... (-36.62 = -38.75 + 2.12)

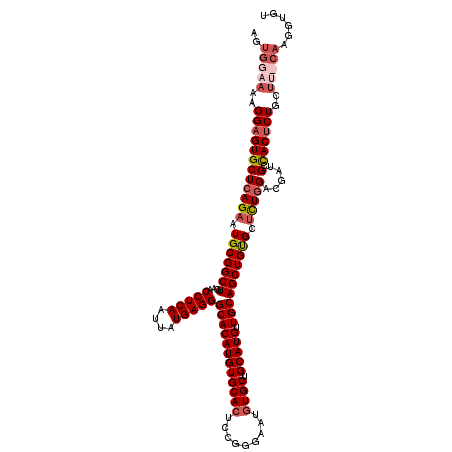
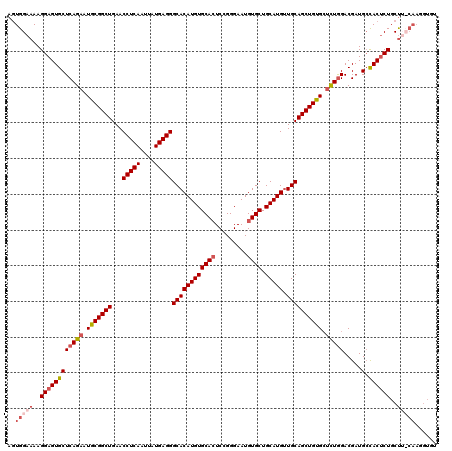
| Location | 17,665,933 – 17,666,053 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.38 |
| Mean single sequence MFE | -37.48 |
| Consensus MFE | -31.36 |
| Energy contribution | -31.68 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.730644 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17665933 120 + 20766785 GCCGUCAUGCAUUAGCUAAUGACAAGCCAUGAAACUAUGGCCAAAUGGGUUGAACAACCUCAGCAGCCGGCCCACUGGGCACCAAAUGAAAUUGUCAUUUCAGAGCGUUGCUGGUGGAGU .((..((.(((...(((((((((((((((((....))))))((..((((((((......))))).....((((...)))).)))..))...))))))))....)))..)))))..))... ( -40.80) >DroSec_CAF1 19368 120 + 1 GCCGUCAUGCAUUAGCUAAUGACAAGCCAUGAAACUAUGGCCAAAUGGGUUGAACAACCUCAGCAGCCGGCCCACUGGGCACCAAAUGAAAUUGUCAUUUCAGAGCGUUGCUGGUGGAGU .((..((.(((...(((((((((((((((((....))))))((..((((((((......))))).....((((...)))).)))..))...))))))))....)))..)))))..))... ( -40.80) >DroEre_CAF1 19742 114 + 1 GCCGUCAUGCAUUAGCUAAUGACAAGCCAUGAAACUAUGGCCAAAUGGGUUGAACAACCUCAGCAGCCGGCCCACUGGGCACCGAAUGAAAUUGUCAUUUCAGAGCGUUGC------AGU .......((((...(((((((((((((((((....))))))((..((((((((......))))).....((((...)))).)))..))...))))))))....)))..)))------).. ( -35.40) >DroYak_CAF1 31438 114 + 1 GCCGUCAUGCAUUAGCUAAUGACAAGCCAUGAAACUAUGGCCAAAUGGGUUGAACAACCUCACCAGCCGGCCCACUGGGCACCAAAUGAAAUUGUCAUUUCAGAGCGUUCC------AGU ((((((((((....))..)))))..((((((....))))))......(((((...........))))))))..((((((.((....((((((....))))))....)))))------))) ( -32.90) >consensus GCCGUCAUGCAUUAGCUAAUGACAAGCCAUGAAACUAUGGCCAAAUGGGUUGAACAACCUCAGCAGCCGGCCCACUGGGCACCAAAUGAAAUUGUCAUUUCAGAGCGUUGC______AGU ........(((...(((((((((((((((((....))))))((..((((((((......))))).....((((...)))).)))..))...))))))))....)))..)))......... (-31.36 = -31.68 + 0.31)

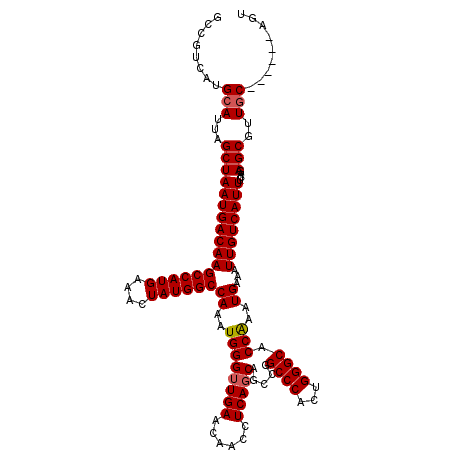
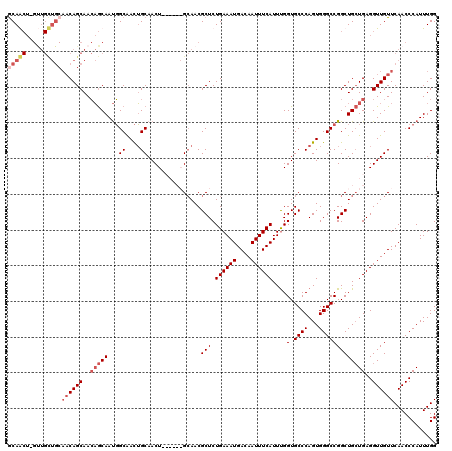
| Location | 17,665,973 – 17,666,093 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.85 |
| Mean single sequence MFE | -38.27 |
| Consensus MFE | -23.51 |
| Energy contribution | -25.82 |
| Covariance contribution | 2.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.61 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.520836 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17665973 120 - 20766785 GCAGCUAGUUGCUGCAACAGCAACAGCAAUGGCAACUGCAACUCCACCAGCAACGCUCUGAAAUGACAAUUUCAUUUGGUGCCCAGUGGGCCGGCUGCUGAGGUUGUUCAACCCAUUUGG (((((.....)))))((((((..(((((.((((.((((......(((((((...))..((((((....))))))..)))))..))))..))))..)))))..))))))............ ( -42.00) >DroSec_CAF1 19408 120 - 1 GCAGCUAGUUGCUGCAACAGCAACAGCAAUGGCAACUGCAACUCCACCAGCAACGCUCUGAAAUGACAAUUUCAUUUGGUGCCCAGUGGGCCGGCUGCUGAGGUUGUUCAACCCAUUUGG (((((.....)))))((((((..(((((.((((.((((......(((((((...))..((((((....))))))..)))))..))))..))))..)))))..))))))............ ( -42.00) >DroEre_CAF1 19782 113 - 1 ACAACU-GCUGUUGCAACAGCAACAGCAAUGGCAACUGCAACU------GCAACGCUCUGAAAUGACAAUUUCAUUCGGUGCCCAGUGGGCCGGCUGCUGAGGUUGUUCAACCCAUUUGG .(((.(-(..((((.((((((..(((((.((((.(((((....------))...((.(((((.(((.....)))))))).))..)))..))))..)))))..)))))))))).)).))). ( -39.10) >DroYak_CAF1 31478 101 - 1 GCAA-------------CAGCAAACACAACUGCGAUGGCAACU------GGAACGCUCUGAAAUGACAAUUUCAUUUGGUGCCCAGUGGGCCGGCUGGUGAGGUUGUUCAACCCAUUUGG (.((-------------((((...(((....((..((((.(((------((..((((.((((((....))))))...)))).)))))..))))))..)))..)))))))........... ( -30.00) >consensus GCAACU_GUUGCUGCAACAGCAACAGCAAUGGCAACUGCAACU______GCAACGCUCUGAAAUGACAAUUUCAUUUGGUGCCCAGUGGGCCGGCUGCUGAGGUUGUUCAACCCAUUUGG (((((.....)))))((((((..(((((...((....))...............(((.((((((....))))))....(.((((...)))))))))))))..))))))............ (-23.51 = -25.82 + 2.31)

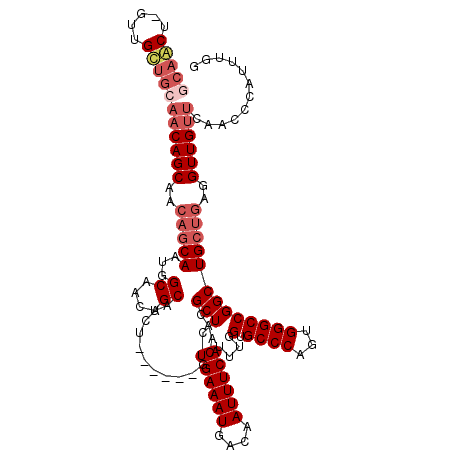
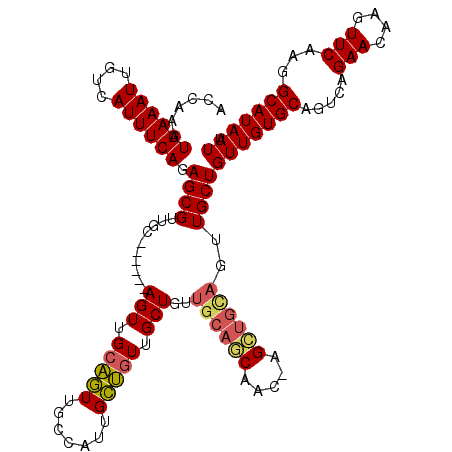
| Location | 17,666,013 – 17,666,133 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.43 |
| Mean single sequence MFE | -33.98 |
| Consensus MFE | -22.49 |
| Energy contribution | -22.93 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.541944 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17666013 120 + 20766785 ACCAAAUGAAAUUGUCAUUUCAGAGCGUUGCUGGUGGAGUUGCAGUUGCCAUUGCUGUUGCUGUUGCAGCAACUAGCUGCAGUUGCUGUUGUGCAGUCAGAACAAGUUCAAGGCAUAAUU .........((((((.((((((.(((...)))..)))))).))))))(((...(((.(((((((.(((((((((......)))))))))...)))).)))....)))....)))...... ( -35.70) >DroSec_CAF1 19448 120 + 1 ACCAAAUGAAAUUGUCAUUUCAGAGCGUUGCUGGUGGAGUUGCAGUUGCCAUUGCUGUUGCUGUUGCAGCAACUAGCUGCAUUUGCUGUUGUGCAGUCAGAACAAGUUCAAGGCAUAAUU .((...((((((....))))))(((((((((((..(.(((.(((((.......))))).))).)..)))))))..(((((((........)))))))........))))..))....... ( -37.20) >DroEre_CAF1 19822 113 + 1 ACCGAAUGAAAUUGUCAUUUCAGAGCGUUGC------AGUUGCAGUUGCCAUUGCUGUUGCUGUUGCAACAGC-AGUUGUAGUUGCUGUUGUGCAGCCAGAACAAGUUCAAGGCAUAAUU ..((..((((((....))))))...))..((------((..(((..(((.(((((((((((....))))))))-))).)))..)))..))))...(((.(((....)))..)))...... ( -39.50) >DroYak_CAF1 31518 101 + 1 ACCAAAUGAAAUUGUCAUUUCAGAGCGUUCC------AGUUGCCAUCGCAGUUGUGUUUGCUG-------------UUGCAGUUGCUGUUGUGCAGUCAGAACAAGUUCAAGGCAUAAUU .((...((((((....))))))((((((((.------..((((.((.(((((.....((((..-------------..))))..))))).))))))...))))..))))..))....... ( -23.50) >consensus ACCAAAUGAAAUUGUCAUUUCAGAGCGUUGC______AGUUGCAGUUGCCAUUGCUGUUGCUGUUGCAGCAAC_AGCUGCAGUUGCUGUUGUGCAGUCAGAACAAGUUCAAGGCAUAAUU ......((((((....)))))).((((..........(((.(((((.......))))).)))..((((((.....))))))..))))(((((((.....(((....)))...))))))). (-22.49 = -22.93 + 0.44)

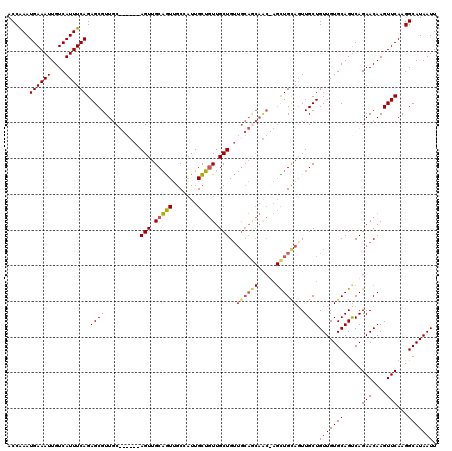
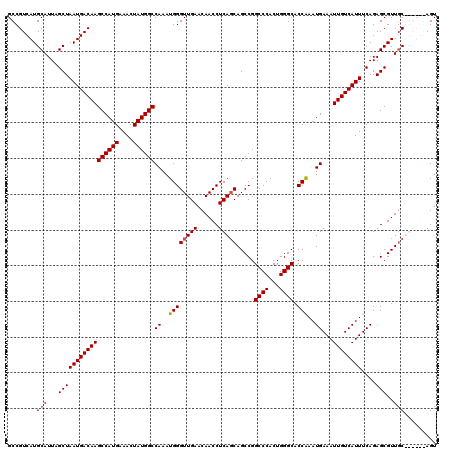
| Location | 17,666,013 – 17,666,133 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.43 |
| Mean single sequence MFE | -28.68 |
| Consensus MFE | -17.67 |
| Energy contribution | -19.55 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.670856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17666013 120 - 20766785 AAUUAUGCCUUGAACUUGUUCUGACUGCACAACAGCAACUGCAGCUAGUUGCUGCAACAGCAACAGCAAUGGCAACUGCAACUCCACCAGCAACGCUCUGAAAUGACAAUUUCAUUUGGU .....(((((((....(((((((..(((......)))..((((((.....)))))).))).)))).))).))))...........((((((...))..((((((....))))))..)))) ( -30.50) >DroSec_CAF1 19448 120 - 1 AAUUAUGCCUUGAACUUGUUCUGACUGCACAACAGCAAAUGCAGCUAGUUGCUGCAACAGCAACAGCAAUGGCAACUGCAACUCCACCAGCAACGCUCUGAAAUGACAAUUUCAUUUGGU .....(((((((....(((((((..(((......)))..((((((.....)))))).))).)))).))).))))...........((((((...))..((((((....))))))..)))) ( -30.70) >DroEre_CAF1 19822 113 - 1 AAUUAUGCCUUGAACUUGUUCUGGCUGCACAACAGCAACUACAACU-GCUGUUGCAACAGCAACAGCAAUGGCAACUGCAACU------GCAACGCUCUGAAAUGACAAUUUCAUUCGGU .....((((......((((....((((.....))))....)))).(-((((((((....)))))))))..))))..(((....------))).((...((((((....))))))..)).. ( -34.30) >DroYak_CAF1 31518 101 - 1 AAUUAUGCCUUGAACUUGUUCUGACUGCACAACAGCAACUGCAA-------------CAGCAAACACAACUGCGAUGGCAACU------GGAACGCUCUGAAAUGACAAUUUCAUUUGGU ......(((........((((((..(((.((.(.((....))..-------------..(((........)))).)))))..)------)))))....((((((....))))))...))) ( -19.20) >consensus AAUUAUGCCUUGAACUUGUUCUGACUGCACAACAGCAACUGCAACU_GUUGCUGCAACAGCAACAGCAAUGGCAACUGCAACU______GCAACGCUCUGAAAUGACAAUUUCAUUUGGU .....(((((((....(((((((..(((......)))..((((((.....)))))).))).)))).))).))))....................(((.((((((....))))))...))) (-17.67 = -19.55 + 1.88)

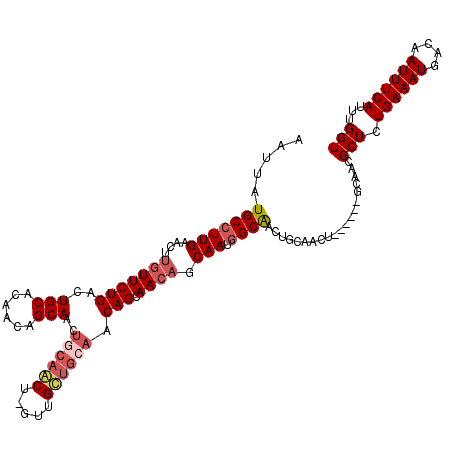
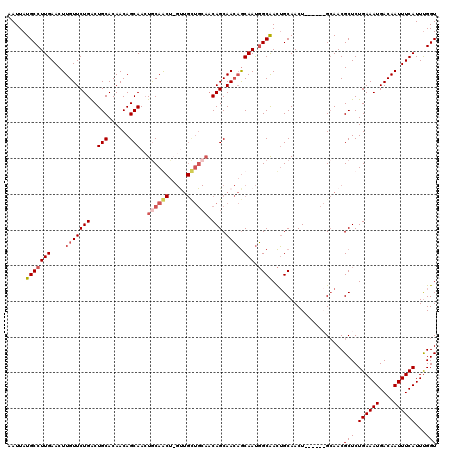
| Location | 17,666,053 – 17,666,173 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.32 |
| Mean single sequence MFE | -44.70 |
| Consensus MFE | -35.21 |
| Energy contribution | -37.52 |
| Covariance contribution | 2.31 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.95 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.986198 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17666053 120 + 20766785 UGCAGUUGCCAUUGCUGUUGCUGUUGCAGCAACUAGCUGCAGUUGCUGUUGUGCAGUCAGAACAAGUUCAAGGCAUAAUUGCUACAAGCAGCAACAUUGGCAGCACUCGUGUGUAAUUGU .((((((((((....(((((((((((((((.....))))).((.((.(((((((.....(((....)))...))))))).)).)).)))))))))).)))).((((....)))))))))) ( -48.20) >DroSec_CAF1 19488 120 + 1 UGCAGUUGCCAUUGCUGUUGCUGUUGCAGCAACUAGCUGCAUUUGCUGUUGUGCAGUCAGAACAAGUUCAAGGCAUAAUUGCUACAUGCAGCAACAUUGGCAGCACUCGUGUGUAAUUGU .((((((((((((((((((..((((((.(((.((..((((((........))))))..))...........((((....))))...))).))))))..)))))))...))).)))))))) ( -46.00) >DroEre_CAF1 19856 119 + 1 UGCAGUUGCCAUUGCUGUUGCUGUUGCAACAGC-AGUUGUAGUUGCUGUUGUGCAGCCAGAACAAGUUCAAGGCAUAAUUGCUACAAGCAGCAACAUUGGCAGCACUCGUGUGUAAUUGU .((((((((((((((((((((....))))))))-)(((((.(((((((((((((((((.(((....)))..))).....))).)).)))))))))....)))))....))).)))))))) ( -50.10) >DroYak_CAF1 31552 107 + 1 UGCCAUCGCAGUUGUGUUUGCUG-------------UUGCAGUUGCUGUUGUGCAGUCAGAACAAGUUCAAGGCAUAAUUGCUACAAGCAGCAACAUUGGCAGCACUCGUGUGUAAUUGU (((((..((((..((....))..-------------)))).(((((((((((.......(((....)))..((((....)))))).)))))))))..)))))((((....))))...... ( -34.50) >consensus UGCAGUUGCCAUUGCUGUUGCUGUUGCAGCAAC_AGCUGCAGUUGCUGUUGUGCAGUCAGAACAAGUUCAAGGCAUAAUUGCUACAAGCAGCAACAUUGGCAGCACUCGUGUGUAAUUGU .((((((((((....(((((((((((((((.....))))))((.((.(((((((.....(((....)))...))))))).)).))..))))))))).)))).((((....)))))))))) (-35.21 = -37.52 + 2.31)

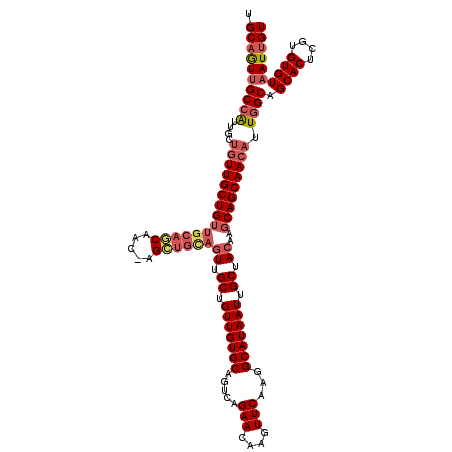
| Location | 17,666,053 – 17,666,173 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.32 |
| Mean single sequence MFE | -38.05 |
| Consensus MFE | -24.34 |
| Energy contribution | -28.77 |
| Covariance contribution | 4.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.66 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798685 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17666053 120 - 20766785 ACAAUUACACACGAGUGCUGCCAAUGUUGCUGCUUGUAGCAAUUAUGCCUUGAACUUGUUCUGACUGCACAACAGCAACUGCAGCUAGUUGCUGCAACAGCAACAGCAAUGGCAACUGCA .......(((....))).(((((.((((((((.((((((((((((.((........((((.((....)).))))((....)).))))))))))))))))))))))....)))))...... ( -41.80) >DroSec_CAF1 19488 120 - 1 ACAAUUACACACGAGUGCUGCCAAUGUUGCUGCAUGUAGCAAUUAUGCCUUGAACUUGUUCUGACUGCACAACAGCAAAUGCAGCUAGUUGCUGCAACAGCAACAGCAAUGGCAACUGCA .......(((....))).(((((.((((((((..(((((((((((.((........((((.((....)).))))((....)).))))))))))))).))))))))....)))))...... ( -39.60) >DroEre_CAF1 19856 119 - 1 ACAAUUACACACGAGUGCUGCCAAUGUUGCUGCUUGUAGCAAUUAUGCCUUGAACUUGUUCUGGCUGCACAACAGCAACUACAACU-GCUGUUGCAACAGCAACAGCAAUGGCAACUGCA .......(((....))).(((((..(((((((.((((.(((.....(((..(((....))).)))))))))))))))))......(-((((((((....))))))))).)))))...... ( -44.60) >DroYak_CAF1 31552 107 - 1 ACAAUUACACACGAGUGCUGCCAAUGUUGCUGCUUGUAGCAAUUAUGCCUUGAACUUGUUCUGACUGCACAACAGCAACUGCAA-------------CAGCAAACACAACUGCGAUGGCA .......(((....)))..((((..(((((((.((((.(((.(((......(((....)))))).)))))))))))))).....-------------..(((........)))..)))). ( -26.20) >consensus ACAAUUACACACGAGUGCUGCCAAUGUUGCUGCUUGUAGCAAUUAUGCCUUGAACUUGUUCUGACUGCACAACAGCAACUGCAACU_GUUGCUGCAACAGCAACAGCAAUGGCAACUGCA .......(((....))).(((((.((((((((.(((((((((((.(((.(((...((((.(.....).))))...)))..)))...)))))))))))))))))))....)))))...... (-24.34 = -28.77 + 4.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:47:56 2006