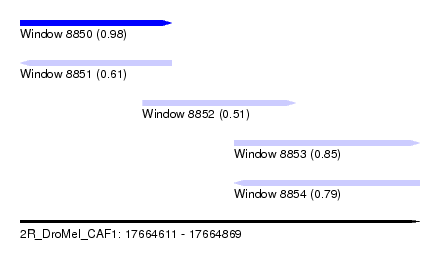
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,664,611 – 17,664,869 |
| Length | 258 |
| Max. P | 0.980636 |

| Location | 17,664,611 – 17,664,709 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.07 |
| Mean single sequence MFE | -33.26 |
| Consensus MFE | -23.61 |
| Energy contribution | -25.43 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17664611 98 + 20766785 AUGUAAAUUGUGCCAAAUCAAUGCCUGGCUCAUAGCCUUGCUGCCGAAUGUGGUAAUUAAAAUCGCUGGGCAGCCAAAG----------------------GAGCGGGCAGUGGGCAUUA .........(((((.......((((((.(((........((((((.(..(((((.......)))))).)))))).....----------------------)))))))))...))))).. ( -33.62) >DroSec_CAF1 18047 98 + 1 AUGUAAAUUGUGCCAAAUCAAUGCCUGGCUCUUAGGCUUGCUGCCGAAUGUGGUAAUUAAAAUCGCUGGGCAGCAAAAG----------------------GAGCGGGCAGCGGGCAUUA .........(((((.......((((((.(((((....((((((((.(..(((((.......)))))).)))))))).))----------------------)))))))))...))))).. ( -37.80) >DroEre_CAF1 18406 97 + 1 AUGUAAAUUGUGGCAAAUCAAUGCCUGGCUCAUAGCCUUGCAGCCGAAUGUGGUAAUUAAAAUCGCUGGGCGGCAAAAA----------------------GAGCAGGCAGCG-GCAUUA ........(((.((.......((((((.(((...((...)).((((...(((((.......)))))....)))).....----------------------))))))))))).-)))... ( -29.71) >DroYak_CAF1 30110 119 + 1 AUGUAAAUUGUGCCAAAUCAAUGCCUGGCUCAUAGCCUUGCUGCCGAAUGUGGUAAUUAAAAUCGCUGGGCAGCAAAAAAAAAAAACAAAACAAAACGAGCGAGCGAUCAGCG-GCAUUA .........(((((.......((((.(((.....)))((((((((.(..(((((.......)))))).)))))))).....................).))).((.....)))-)))).. ( -31.90) >consensus AUGUAAAUUGUGCCAAAUCAAUGCCUGGCUCAUAGCCUUGCUGCCGAAUGUGGUAAUUAAAAUCGCUGGGCAGCAAAAA______________________GAGCGGGCAGCG_GCAUUA .........(((((.......((((((((.....)))((((((((.(..(((((.......)))))).)))))))).............................)))))...))))).. (-23.61 = -25.43 + 1.81)


| Location | 17,664,611 – 17,664,709 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.07 |
| Mean single sequence MFE | -26.84 |
| Consensus MFE | -17.38 |
| Energy contribution | -18.62 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.609181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17664611 98 - 20766785 UAAUGCCCACUGCCCGCUC----------------------CUUUGGCUGCCCAGCGAUUUUAAUUACCACAUUCGGCAGCAAGGCUAUGAGCCAGGCAUUGAUUUGGCACAAUUUACAU ...((((((.((((.((((----------------------(((((.(((((.......................))))))))))....))))..)))).))....)))).......... ( -26.50) >DroSec_CAF1 18047 98 - 1 UAAUGCCCGCUGCCCGCUC----------------------CUUUUGCUGCCCAGCGAUUUUAAUUACCACAUUCGGCAGCAAGCCUAAGAGCCAGGCAUUGAUUUGGCACAAUUUACAU ...((((((.((((.((((----------------------..(((((((((.......................))))))))).....))))..)))).))....)))).......... ( -26.70) >DroEre_CAF1 18406 97 - 1 UAAUGC-CGCUGCCUGCUC----------------------UUUUUGCCGCCCAGCGAUUUUAAUUACCACAUUCGGCUGCAAGGCUAUGAGCCAGGCAUUGAUUUGCCACAAUUUACAU ......-((.(((((((((----------------------.....(((...((((((...............)).))))...)))...))).)))))).)).................. ( -24.86) >DroYak_CAF1 30110 119 - 1 UAAUGC-CGCUGAUCGCUCGCUCGUUUUGUUUUGUUUUUUUUUUUUGCUGCCCAGCGAUUUUAAUUACCACAUUCGGCAGCAAGGCUAUGAGCCAGGCAUUGAUUUGGCACAAUUUACAU ((((((-(((.....))..((((((...((((((((........(((((....))))).........((......)).)))))))).))))))..))))))).................. ( -29.30) >consensus UAAUGC_CGCUGCCCGCUC______________________CUUUUGCUGCCCAGCGAUUUUAAUUACCACAUUCGGCAGCAAGGCUAUGAGCCAGGCAUUGAUUUGGCACAAUUUACAU (((((((.....................................((((((((.......................))))))))(((.....))).))))))).................. (-17.38 = -18.62 + 1.25)

| Location | 17,664,690 – 17,664,789 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.35 |
| Mean single sequence MFE | -32.83 |
| Consensus MFE | -28.31 |
| Energy contribution | -28.62 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.86 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.510493 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17664690 99 + 20766785 ---------------------GAGCGGGCAGUGGGCAUUAGUAUGCAACUCUCACUCGGCGACAGCGGCCGCAACUGUUGGCCAUAUAAGCGGACUGAAAAGAAAGGCCAACGCUCAGUG ---------------------(((((((((((..((((....)))).))).(((.((.((......((((((....).)))))......)).)).)))........)))..))))).... ( -32.90) >DroSec_CAF1 18126 99 + 1 ---------------------GAGCGGGCAGCGGGCAUUAGUAUGCAACUCUCACUCGGCGACAGCGGCCGCAACUGUUGGCCAUAUAAGCGGACUGAAAAGAAAGGCCAACGCUCAGUG ---------------------((((((.((((((((((....))))..........((((.......))))...)))))).))........((.((........)).))...)))).... ( -31.90) >DroEre_CAF1 18485 98 + 1 ---------------------GAGCAGGCAGCG-GCAUUAGUAUGCAACUCUCACUCGGAGACAGCGGCCGCAACUGUUGGCCAUAUAAGCGGACUGAAAAGAAAGGCCAACGCUCAGUG ---------------------((((.(((.((.-((((....))))...((((.....))))..)).)))......(((((((......................))))))))))).... ( -33.25) >DroYak_CAF1 30190 119 + 1 AAAAAACAAAACAAAACGAGCGAGCGAUCAGCG-GCAUUAGUAUGCAACUCUCACUCGGAGACAGCGGCCGCAACUGUUGGCCAUAUAAGCGGACUGAAAAGAAAGGCCAACGCUCAGUG .................((((..(((..(.((.-((((....))))...((((.....))))..)))..)))....(((((((......................))))))))))).... ( -33.25) >consensus _____________________GAGCGGGCAGCG_GCAUUAGUAUGCAACUCUCACUCGGAGACAGCGGCCGCAACUGUUGGCCAUAUAAGCGGACUGAAAAGAAAGGCCAACGCUCAGUG .....................((((.(((.((..((((....))))...........(....).)).)))......(((((((......................))))))))))).... (-28.31 = -28.62 + 0.31)

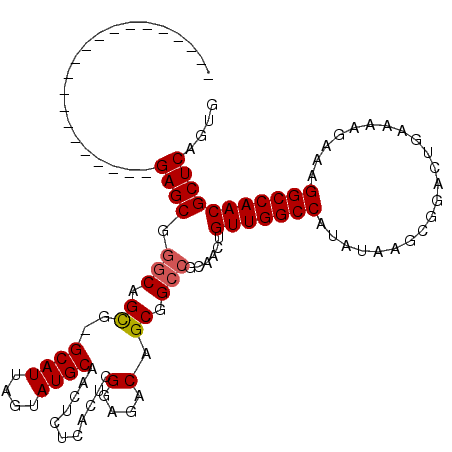
| Location | 17,664,749 – 17,664,869 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -34.06 |
| Consensus MFE | -32.96 |
| Energy contribution | -32.96 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.854448 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17664749 120 + 20766785 GCCAUAUAAGCGGACUGAAAAGAAAGGCCAACGCUCAGUGCUCAACAAAACAAUUUACCAAGUUUGGCCUAAACGGCCAGUGACAGUCCGGCGACAACAACAAUUGCAACUGUUACGGCA (((......((.(.((....))..(((((((.(((..(((...............)))..))))))))))...).))..((((((((...((((.........)))).))))))))))). ( -34.06) >DroSec_CAF1 18185 120 + 1 GCCAUAUAAGCGGACUGAAAAGAAAGGCCAACGCUCAGUGCUCAACAAAACAAUUUACCAAGUUUGGCCUAAACGGCCAGUGACAGUCCGGCGACAACAACAAUUGCAACUGUUACGGCA (((......((.(.((....))..(((((((.(((..(((...............)))..))))))))))...).))..((((((((...((((.........)))).))))))))))). ( -34.06) >DroEre_CAF1 18543 120 + 1 GCCAUAUAAGCGGACUGAAAAGAAAGGCCAACGCUCAGUGCUCAACAAAACAAUUUACCAAGUUUGGCCUAAACGGCCAGUGACAGUCCGGCGACAACAACAAUUGCAACUGUUACGGCA (((......((.(.((....))..(((((((.(((..(((...............)))..))))))))))...).))..((((((((...((((.........)))).))))))))))). ( -34.06) >DroYak_CAF1 30269 120 + 1 GCCAUAUAAGCGGACUGAAAAGAAAGGCCAACGCUCAGUGCUCAACAAAACAAUUUACCAAGUUUGGCCUAAACGGCCAGUGACAGUCCGGCGACAACAACAAUUGCAACUGUUACGGCA (((......((.(.((....))..(((((((.(((..(((...............)))..))))))))))...).))..((((((((...((((.........)))).))))))))))). ( -34.06) >consensus GCCAUAUAAGCGGACUGAAAAGAAAGGCCAACGCUCAGUGCUCAACAAAACAAUUUACCAAGUUUGGCCUAAACGGCCAGUGACAGUCCGGCGACAACAACAAUUGCAACUGUUACGGCA (((........((.(((.......(((((((.(((..(((...............)))..))))))))))...))))).((((((((...((((.........)))).))))))))))). (-32.96 = -32.96 + 0.00)



| Location | 17,664,749 – 17,664,869 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -39.62 |
| Consensus MFE | -39.62 |
| Energy contribution | -39.62 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.23 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.785168 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17664749 120 - 20766785 UGCCGUAACAGUUGCAAUUGUUGUUGUCGCCGGACUGUCACUGGCCGUUUAGGCCAAACUUGGUAAAUUGUUUUGUUGAGCACUGAGCGUUGGCCUUUCUUUUCAGUCCGCUUAUAUGGC .((((((...((.(((((....))))).))(((((((.....((((((((((((((....))))....(((((....)))))))))))...))))........)))))))....)))))) ( -39.62) >DroSec_CAF1 18185 120 - 1 UGCCGUAACAGUUGCAAUUGUUGUUGUCGCCGGACUGUCACUGGCCGUUUAGGCCAAACUUGGUAAAUUGUUUUGUUGAGCACUGAGCGUUGGCCUUUCUUUUCAGUCCGCUUAUAUGGC .((((((...((.(((((....))))).))(((((((.....((((((((((((((....))))....(((((....)))))))))))...))))........)))))))....)))))) ( -39.62) >DroEre_CAF1 18543 120 - 1 UGCCGUAACAGUUGCAAUUGUUGUUGUCGCCGGACUGUCACUGGCCGUUUAGGCCAAACUUGGUAAAUUGUUUUGUUGAGCACUGAGCGUUGGCCUUUCUUUUCAGUCCGCUUAUAUGGC .((((((...((.(((((....))))).))(((((((.....((((((((((((((....))))....(((((....)))))))))))...))))........)))))))....)))))) ( -39.62) >DroYak_CAF1 30269 120 - 1 UGCCGUAACAGUUGCAAUUGUUGUUGUCGCCGGACUGUCACUGGCCGUUUAGGCCAAACUUGGUAAAUUGUUUUGUUGAGCACUGAGCGUUGGCCUUUCUUUUCAGUCCGCUUAUAUGGC .((((((...((.(((((....))))).))(((((((.....((((((((((((((....))))....(((((....)))))))))))...))))........)))))))....)))))) ( -39.62) >consensus UGCCGUAACAGUUGCAAUUGUUGUUGUCGCCGGACUGUCACUGGCCGUUUAGGCCAAACUUGGUAAAUUGUUUUGUUGAGCACUGAGCGUUGGCCUUUCUUUUCAGUCCGCUUAUAUGGC .((((((...((.(((((....))))).))(((((((.....((((((((((((((....))))....(((((....)))))))))))...))))........)))))))....)))))) (-39.62 = -39.62 + 0.00)

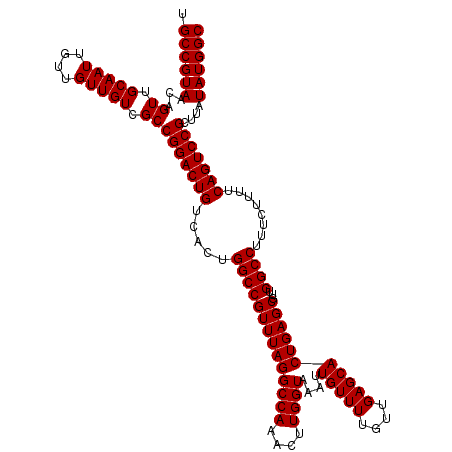
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:47:43 2006