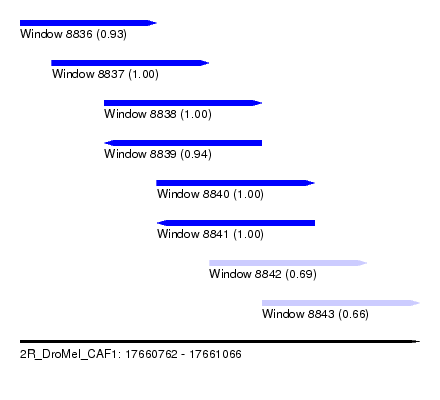
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,660,762 – 17,661,066 |
| Length | 304 |
| Max. P | 0.999953 |

| Location | 17,660,762 – 17,660,866 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.31 |
| Mean single sequence MFE | -45.12 |
| Consensus MFE | -33.79 |
| Energy contribution | -33.99 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934999 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17660762 104 + 20766785 GUG----CAGUGGCGGUG------------GCAGCAGCACCAACUUGAUGCAUCAUCCUCGCACCGGCGAGGGAUUCAUAUCUCCAUCGGAGGCUGUAAUCUGCUGGCUACUGUGCGUUG (..----(((((((((..------------(..(((((.........(((.(((..((((((....))))))))).)))..((((...)))))))))...)..)).)))))))..).... ( -44.10) >DroSec_CAF1 14216 106 + 1 GUGC--GCAGUGGCAGUG------------GCAGCAGCACCAACUUGAUGCAUCAUCCUCGCAUCGGCGAGGGAUUCAUAUCUCCAUAGGAGGCUGUAAUCUGCUGGCUACUGUGCGUUG ..((--((((((((((..------------(..(((((.........(((.(((..((((((....))))))))).)))..((((...)))))))))...)..)).)))))))))).... ( -47.20) >DroSim_CAF1 20230 106 + 1 GUGC--GCAGUGGCAGUG------------GCAGCAGCACCAACUUGAUGCAUCAUCCUCGCAUCGGCGAGGGAUUCAUAUCUCCAUCGGAGGCUGUAAUCUGCUGGCUACUGUACGUUG ..(.--.(((((((((..------------(..(((((.........(((.(((..((((((....))))))))).)))..((((...)))))))))...)..)).)))))))..).... ( -41.80) >DroEre_CAF1 14554 104 + 1 GU----GCAGUGGCAGUG------------GCAGCAGCACCAACUUGAUGCAUCAUCCUCGCAUCGGCGGGGGAUUCAUAUCUCCAUCGGAGGCUGUAAUCUCCUGGCUACUGUGCGUUG (.----.(((((((((..------------(..(((((.((...(((((((.........)))))))..((((((....))))))...))..)))))...)..)).)))))))..).... ( -41.60) >DroYak_CAF1 26044 120 + 1 GUAUGUCCAGUGGCAGUGGCGGUGGCAGCAGCAGCAGCAUCAACUUGAUGCAUCAUCCUCGCAUCGGCGGGGGAUUCAUAUCUCCAUCGGAGGCUGUAAUCUGCUGGCUACUGUGCGUUG ..(((..(((((((...(((((..(((((.......(((((.....)))))....(((((((....)))))))........((((...)))))))))...))))).)))))))..))).. ( -50.90) >consensus GUG___GCAGUGGCAGUG____________GCAGCAGCACCAACUUGAUGCAUCAUCCUCGCAUCGGCGAGGGAUUCAUAUCUCCAUCGGAGGCUGUAAUCUGCUGGCUACUGUGCGUUG ......((((((((((((............(((((.(((.(.....).)))....(((((((....)))))))........((((...)))))))))....)))).))))))))...... (-33.79 = -33.99 + 0.20)

| Location | 17,660,786 – 17,660,906 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.08 |
| Mean single sequence MFE | -45.48 |
| Consensus MFE | -43.28 |
| Energy contribution | -43.44 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.36 |
| Structure conservation index | 0.95 |
| SVM decision value | 4.82 |
| SVM RNA-class probability | 0.999953 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17660786 120 + 20766785 CAACUUGAUGCAUCAUCCUCGCACCGGCGAGGGAUUCAUAUCUCCAUCGGAGGCUGUAAUCUGCUGGCUACUGUGCGUUGUCACAGCCAGCUCCUGCGGCCUCUGUUCUGCCUAGUCAUC .....(((((((...(((((((....)))))))..............(((((((((((....(((((((...(((......))))))))))...)))))))))))...)))...)))).. ( -46.90) >DroSec_CAF1 14242 120 + 1 CAACUUGAUGCAUCAUCCUCGCAUCGGCGAGGGAUUCAUAUCUCCAUAGGAGGCUGUAAUCUGCUGGCUACUGUGCGUUGUCACAGCCAGCUCCUGCGGCCUCUGUUCUGCAUAGUCAUC .......(((((...(((((((....)))))))...............((((((((((....(((((((...(((......))))))))))...))))))))))....)))))....... ( -46.60) >DroSim_CAF1 20256 120 + 1 CAACUUGAUGCAUCAUCCUCGCAUCGGCGAGGGAUUCAUAUCUCCAUCGGAGGCUGUAAUCUGCUGGCUACUGUACGUUGUCACAGCCAGCUCCUGCGGCCUCUGUUCUGCCUAGUCAUC .....(((((((...(((((((....)))))))..............(((((((((((....(((((((((........))...)))))))...)))))))))))...)))...)))).. ( -45.60) >DroEre_CAF1 14578 120 + 1 CAACUUGAUGCAUCAUCCUCGCAUCGGCGGGGGAUUCAUAUCUCCAUCGGAGGCUGUAAUCUCCUGGCUACUGUGCGUUGUCACAGCCAGCUCCUGCGGCCUCUAUUCUGCAUAGUCAUC ......(((((.........)))))(((.((((((....))))))...((((((((((.....((((((...(((......)))))))))....))))))))))..........)))... ( -42.30) >DroYak_CAF1 26084 120 + 1 CAACUUGAUGCAUCAUCCUCGCAUCGGCGGGGGAUUCAUAUCUCCAUCGGAGGCUGUAAUCUGCUGGCUACUGUGCGUUGUCACAGCCAGCUCCUGCGGCCUCUCUUCUGCAUAGUCAUC ......(((((.........)))))(((.((((((....))))))...((((((((((....(((((((...(((......))))))))))...))))))))))..........)))... ( -46.00) >consensus CAACUUGAUGCAUCAUCCUCGCAUCGGCGAGGGAUUCAUAUCUCCAUCGGAGGCUGUAAUCUGCUGGCUACUGUGCGUUGUCACAGCCAGCUCCUGCGGCCUCUGUUCUGCAUAGUCAUC .....(((((((...(((((((....)))))))...............((((((((((....(((((((...(((......))))))))))...))))))))))....)))...)))).. (-43.28 = -43.44 + 0.16)

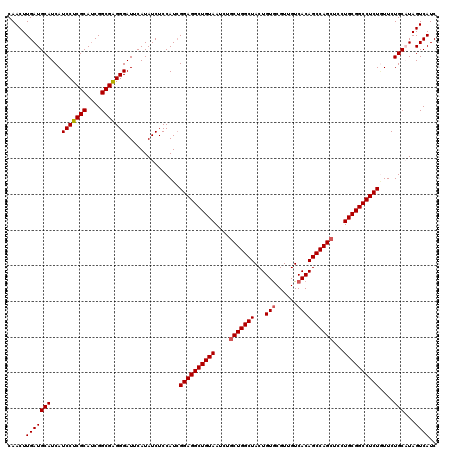
| Location | 17,660,826 – 17,660,946 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.92 |
| Mean single sequence MFE | -39.66 |
| Consensus MFE | -38.32 |
| Energy contribution | -39.12 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.72 |
| Structure conservation index | 0.97 |
| SVM decision value | 3.79 |
| SVM RNA-class probability | 0.999615 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17660826 120 + 20766785 UCUCCAUCGGAGGCUGUAAUCUGCUGGCUACUGUGCGUUGUCACAGCCAGCUCCUGCGGCCUCUGUUCUGCCUAGUCAUCCUUAAUUAUUAUGAUUAUCGAUGUUUUAUGCUCUAUAAUU .......(((((((((((....(((((((...(((......))))))))))...)))))))))))....((.(((.((((..(((((.....)))))..))))..))).))......... ( -39.20) >DroSec_CAF1 14282 120 + 1 UCUCCAUAGGAGGCUGUAAUCUGCUGGCUACUGUGCGUUGUCACAGCCAGCUCCUGCGGCCUCUGUUCUGCAUAGUCAUCCUUAAUUAUUAUGAUUAUCGAUGUUUUAUGCUCUAUAAUU .....(((((((((((((....(((((((...(((......))))))))))...)))))))))......((((((.((((..(((((.....)))))..))))..)))))).)))).... ( -42.30) >DroSim_CAF1 20296 120 + 1 UCUCCAUCGGAGGCUGUAAUCUGCUGGCUACUGUACGUUGUCACAGCCAGCUCCUGCGGCCUCUGUUCUGCCUAGUCAUCCUUAAUUAUUAUGAUUAUCGAUGUUUUAUGCUCUAUAAUU .......(((((((((((....(((((((((........))...)))))))...)))))))))))....((.(((.((((..(((((.....)))))..))))..))).))......... ( -37.90) >DroEre_CAF1 14618 120 + 1 UCUCCAUCGGAGGCUGUAAUCUCCUGGCUACUGUGCGUUGUCACAGCCAGCUCCUGCGGCCUCUAUUCUGCAUAGUCAUCCUUAAUUAUUAUGAUUAUCGAUGUUUUAUGCUCUAUAAUU ........((((((((((.....((((((...(((......)))))))))....)))))))))).....((((((.((((..(((((.....)))))..))))..))))))......... ( -37.60) >DroYak_CAF1 26124 120 + 1 UCUCCAUCGGAGGCUGUAAUCUGCUGGCUACUGUGCGUUGUCACAGCCAGCUCCUGCGGCCUCUCUUCUGCAUAGUCAUCCUUAAUUAUUAUGAUUAUCGAUGUUUUAUGCUCUAUAAUU ........((((((((((....(((((((...(((......))))))))))...)))))))))).....((((((.((((..(((((.....)))))..))))..))))))......... ( -41.30) >consensus UCUCCAUCGGAGGCUGUAAUCUGCUGGCUACUGUGCGUUGUCACAGCCAGCUCCUGCGGCCUCUGUUCUGCAUAGUCAUCCUUAAUUAUUAUGAUUAUCGAUGUUUUAUGCUCUAUAAUU ........((((((((((....(((((((...(((......))))))))))...)))))))))).....((((((.((((..(((((.....)))))..))))..))))))......... (-38.32 = -39.12 + 0.80)

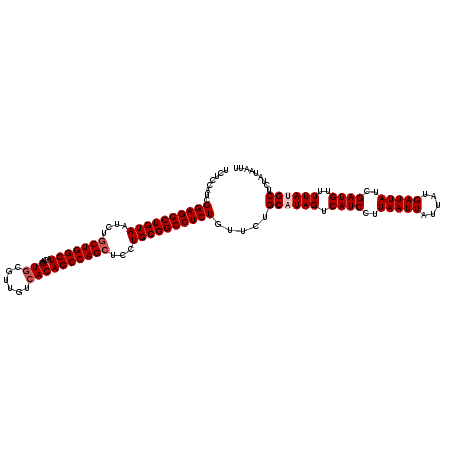
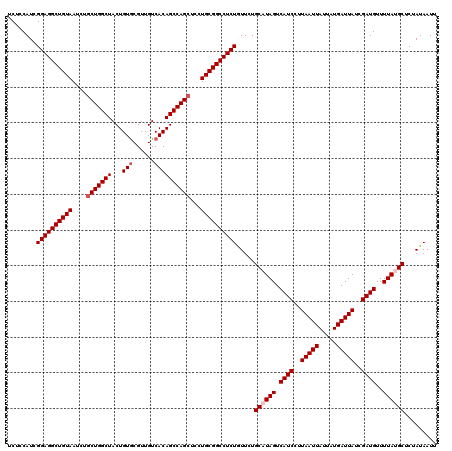
| Location | 17,660,826 – 17,660,946 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.92 |
| Mean single sequence MFE | -30.02 |
| Consensus MFE | -27.50 |
| Energy contribution | -27.94 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938158 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17660826 120 - 20766785 AAUUAUAGAGCAUAAAACAUCGAUAAUCAUAAUAAUUAAGGAUGACUAGGCAGAACAGAGGCCGCAGGAGCUGGCUGUGACAACGCACAGUAGCCAGCAGAUUACAGCCUCCGAUGGAGA .................(((((....((((..(.....)..))))............(((((.(.((..((((((((((......)))...)))))))...)).).)))))))))).... ( -29.20) >DroSec_CAF1 14282 120 - 1 AAUUAUAGAGCAUAAAACAUCGAUAAUCAUAAUAAUUAAGGAUGACUAUGCAGAACAGAGGCCGCAGGAGCUGGCUGUGACAACGCACAGUAGCCAGCAGAUUACAGCCUCCUAUGGAGA ..((((((.(((((...((((..((((.......))))..))))..)))))......(((((.(.((..((((((((((......)))...)))))))...)).).)))))))))))... ( -33.90) >DroSim_CAF1 20296 120 - 1 AAUUAUAGAGCAUAAAACAUCGAUAAUCAUAAUAAUUAAGGAUGACUAGGCAGAACAGAGGCCGCAGGAGCUGGCUGUGACAACGUACAGUAGCCAGCAGAUUACAGCCUCCGAUGGAGA .................(((((....((((..(.....)..))))............(((((.(.((..((((((((((........)).))))))))...)).).)))))))))).... ( -28.90) >DroEre_CAF1 14618 120 - 1 AAUUAUAGAGCAUAAAACAUCGAUAAUCAUAAUAAUUAAGGAUGACUAUGCAGAAUAGAGGCCGCAGGAGCUGGCUGUGACAACGCACAGUAGCCAGGAGAUUACAGCCUCCGAUGGAGA .................((((..................((...(((.(((........((((((....)).))))((....))))).)))..)).((((........)))))))).... ( -27.20) >DroYak_CAF1 26124 120 - 1 AAUUAUAGAGCAUAAAACAUCGAUAAUCAUAAUAAUUAAGGAUGACUAUGCAGAAGAGAGGCCGCAGGAGCUGGCUGUGACAACGCACAGUAGCCAGCAGAUUACAGCCUCCGAUGGAGA .........(((((...((((..((((.......))))..))))..)))))....(.(((((.(.((..((((((((((......)))...)))))))...)).).))))))........ ( -30.90) >consensus AAUUAUAGAGCAUAAAACAUCGAUAAUCAUAAUAAUUAAGGAUGACUAUGCAGAACAGAGGCCGCAGGAGCUGGCUGUGACAACGCACAGUAGCCAGCAGAUUACAGCCUCCGAUGGAGA .........(((((...((((..((((.......))))..))))..)))))......(((((.(.((..((((((((((......)))...)))))))...)).).)))))......... (-27.50 = -27.94 + 0.44)

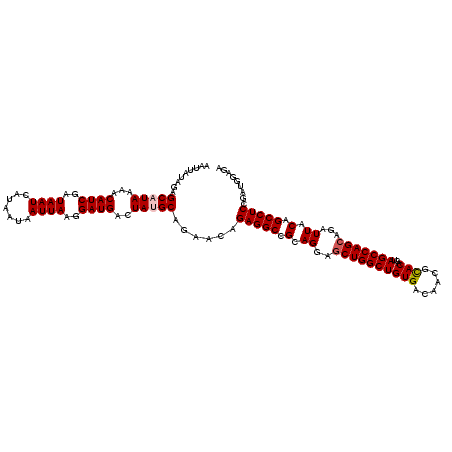
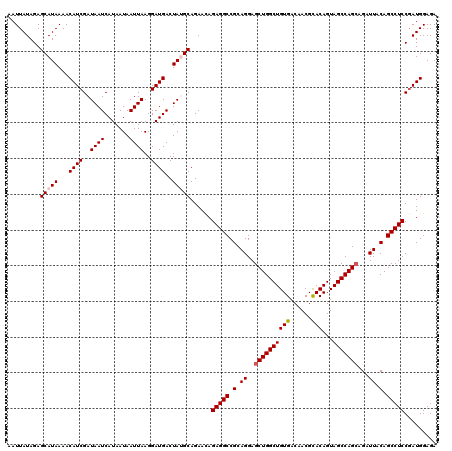
| Location | 17,660,866 – 17,660,986 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.08 |
| Mean single sequence MFE | -41.93 |
| Consensus MFE | -40.74 |
| Energy contribution | -41.42 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.71 |
| Structure conservation index | 0.97 |
| SVM decision value | 4.58 |
| SVM RNA-class probability | 0.999923 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17660866 120 + 20766785 UCACAGCCAGCUCCUGCGGCCUCUGUUCUGCCUAGUCAUCCUUAAUUAUUAUGAUUAUCGAUGUUUUAUGCUCUAUAAUUGCCGCUGGCGGUGCGGAGUUGGCAGUGACUUGGAGUUUUU ((((.(((((((((.(((((...(((...((.(((.((((..(((((.....)))))..))))..))).))...)))...)))))..((...))))))))))).))))............ ( -39.90) >DroSec_CAF1 14322 120 + 1 UCACAGCCAGCUCCUGCGGCCUCUGUUCUGCAUAGUCAUCCUUAAUUAUUAUGAUUAUCGAUGUUUUAUGCUCUAUAAUUGCCGAUGGCGGUGCGGAGUUGGCAGUGACUUGGAGUUUUU ((((.(((((((((.(((.((..(((...((((((.((((..(((((.....)))))..))))..))))))...)))...(((...))))))))))))))))).))))............ ( -42.30) >DroSim_CAF1 20336 120 + 1 UCACAGCCAGCUCCUGCGGCCUCUGUUCUGCCUAGUCAUCCUUAAUUAUUAUGAUUAUCGAUGUUUUAUGCUCUAUAAUUGCCGCUGGCGGUGCGGAGUUGGCAGUGACUUGGAGUUUUU ((((.(((((((((.(((((...(((...((.(((.((((..(((((.....)))))..))))..))).))...)))...)))))..((...))))))))))).))))............ ( -39.90) >DroEre_CAF1 14658 120 + 1 UCACAGCCAGCUCCUGCGGCCUCUAUUCUGCAUAGUCAUCCUUAAUUAUUAUGAUUAUCGAUGUUUUAUGCUCUAUAAUUGCCGCUGGCGGUGCGGAGUUGGCAGUGACUUGGACUUUUU ((((.(((((((((.(((((...(((...((((((.((((..(((((.....)))))..))))..))))))...)))...)))))..((...))))))))))).))))............ ( -44.20) >DroYak_CAF1 26164 120 + 1 UCACAGCCAGCUCCUGCGGCCUCUCUUCUGCAUAGUCAUCCUUAAUUAUUAUGAUUAUCGAUGUUUUAUGCUCUAUAAUUGCCGCUGGCGGUGCGGAGUUGGCAGUGACUUGGACUUUUU ((((.(((((((((.(((((.........((((((.((((..(((((.....)))))..))))..)))))).........)))))..((...))))))))))).))))............ ( -43.37) >consensus UCACAGCCAGCUCCUGCGGCCUCUGUUCUGCAUAGUCAUCCUUAAUUAUUAUGAUUAUCGAUGUUUUAUGCUCUAUAAUUGCCGCUGGCGGUGCGGAGUUGGCAGUGACUUGGAGUUUUU ((((.(((((((((.(((((...(((...((((((.((((..(((((.....)))))..))))..))))))...)))...)))))..((...))))))))))).))))............ (-40.74 = -41.42 + 0.68)


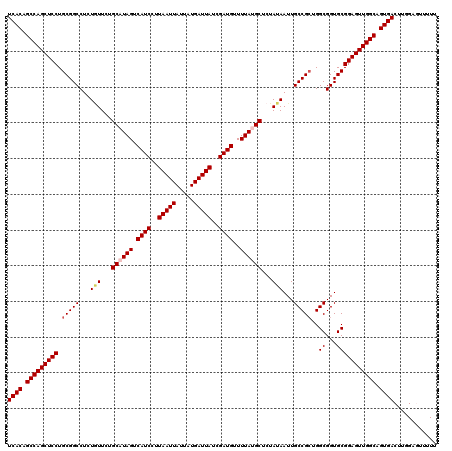
| Location | 17,660,866 – 17,660,986 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.08 |
| Mean single sequence MFE | -34.60 |
| Consensus MFE | -33.23 |
| Energy contribution | -33.83 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.96 |
| SVM decision value | 4.31 |
| SVM RNA-class probability | 0.999866 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17660866 120 - 20766785 AAAAACUCCAAGUCACUGCCAACUCCGCACCGCCAGCGGCAAUUAUAGAGCAUAAAACAUCGAUAAUCAUAAUAAUUAAGGAUGACUAGGCAGAACAGAGGCCGCAGGAGCUGGCUGUGA ............((((.((((.((((((...))..(((((.........((......((((..((((.......))))..)))).....)).........))))).)))).)))).)))) ( -32.77) >DroSec_CAF1 14322 120 - 1 AAAAACUCCAAGUCACUGCCAACUCCGCACCGCCAUCGGCAAUUAUAGAGCAUAAAACAUCGAUAAUCAUAAUAAUUAAGGAUGACUAUGCAGAACAGAGGCCGCAGGAGCUGGCUGUGA ............((((.((((.((((((.(((((...))).........(((((...((((..((((.......))))..))))..)))))........))..)).)))).)))).)))) ( -33.60) >DroSim_CAF1 20336 120 - 1 AAAAACUCCAAGUCACUGCCAACUCCGCACCGCCAGCGGCAAUUAUAGAGCAUAAAACAUCGAUAAUCAUAAUAAUUAAGGAUGACUAGGCAGAACAGAGGCCGCAGGAGCUGGCUGUGA ............((((.((((.((((((...))..(((((.........((......((((..((((.......))))..)))).....)).........))))).)))).)))).)))) ( -32.77) >DroEre_CAF1 14658 120 - 1 AAAAAGUCCAAGUCACUGCCAACUCCGCACCGCCAGCGGCAAUUAUAGAGCAUAAAACAUCGAUAAUCAUAAUAAUUAAGGAUGACUAUGCAGAAUAGAGGCCGCAGGAGCUGGCUGUGA ............((((.((((.((((((...))..(((((..((((...(((((...((((..((((.......))))..))))..)))))...))))..))))).)))).)))).)))) ( -37.60) >DroYak_CAF1 26164 120 - 1 AAAAAGUCCAAGUCACUGCCAACUCCGCACCGCCAGCGGCAAUUAUAGAGCAUAAAACAUCGAUAAUCAUAAUAAUUAAGGAUGACUAUGCAGAAGAGAGGCCGCAGGAGCUGGCUGUGA ............((((.((((.((((((...))..(((((.........(((((...((((..((((.......))))..))))..))))).........))))).)))).)))).)))) ( -36.27) >consensus AAAAACUCCAAGUCACUGCCAACUCCGCACCGCCAGCGGCAAUUAUAGAGCAUAAAACAUCGAUAAUCAUAAUAAUUAAGGAUGACUAUGCAGAACAGAGGCCGCAGGAGCUGGCUGUGA ............((((.((((.((((((...))..(((((.........(((((...((((..((((.......))))..))))..))))).........))))).)))).)))).)))) (-33.23 = -33.83 + 0.60)

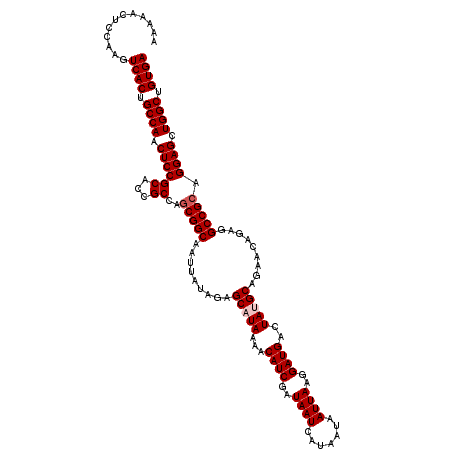
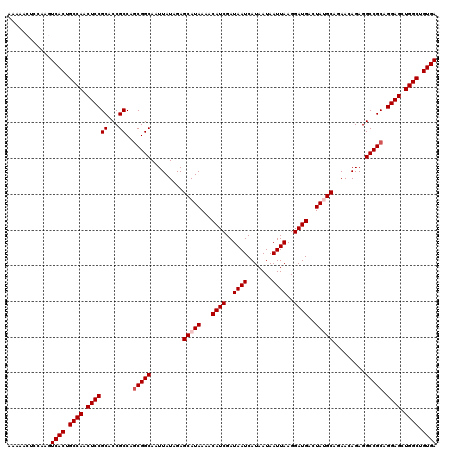
| Location | 17,660,906 – 17,661,026 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.00 |
| Mean single sequence MFE | -34.14 |
| Consensus MFE | -33.42 |
| Energy contribution | -34.02 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.690800 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17660906 120 + 20766785 CUUAAUUAUUAUGAUUAUCGAUGUUUUAUGCUCUAUAAUUGCCGCUGGCGGUGCGGAGUUGGCAGUGACUUGGAGUUUUUGGGCGUCUCCCAGCCCAGCCUCCAAAAUUGCUGUCUUCAU ..........((((...............(((((...((((((...))))))..))))).(((((..(.((((((...((((((........)))))).))))))..)..))))).)))) ( -37.80) >DroSec_CAF1 14362 120 + 1 CUUAAUUAUUAUGAUUAUCGAUGUUUUAUGCUCUAUAAUUGCCGAUGGCGGUGCGGAGUUGGCAGUGACUUGGAGUUUUUGGGCGUCUCCCAGUCCAGCCUCCAAAAUUGCUGUCUUCAU ..........((((...............(((((...((((((...))))))..))))).(((((..(.((((((...((((((........)))))).))))))..)..))))).)))) ( -35.10) >DroSim_CAF1 20376 120 + 1 CUUAAUUAUUAUGAUUAUCGAUGUUUUAUGCUCUAUAAUUGCCGCUGGCGGUGCGGAGUUGGCAGUGACUUGGAGUUUUUGGGCGUCUCCCAGUCCAGCCUCCAAAAUUGCUGUCUUCAU ..........((((...............(((((...((((((...))))))..))))).(((((..(.((((((...((((((........)))))).))))))..)..))))).)))) ( -35.10) >DroEre_CAF1 14698 120 + 1 CUUAAUUAUUAUGAUUAUCGAUGUUUUAUGCUCUAUAAUUGCCGCUGGCGGUGCGGAGUUGGCAGUGACUUGGACUUUUUGGGCGUCUCCCGGCCCAGCCCCCAAAAUUGCUGUCUUCAU ..........((((...............(((((...((((((...))))))..))))).(((((..(.((((.....((((((........))))))...))))..)..))))).)))) ( -32.10) >DroYak_CAF1 26204 120 + 1 CUUAAUUAUUAUGAUUAUCGAUGUUUUAUGCUCUAUAAUUGCCGCUGGCGGUGCGGAGUUGGCAGUGACUUGGACUUUUUGGGCGUCUCCCGGCCCGCCCCCCAAAAUUGCUGUCUUCAU ..........((((...............(((((...((((((...))))))..))))).(((((..(.((((......(((((........)))))....))))..)..))))).)))) ( -30.60) >consensus CUUAAUUAUUAUGAUUAUCGAUGUUUUAUGCUCUAUAAUUGCCGCUGGCGGUGCGGAGUUGGCAGUGACUUGGAGUUUUUGGGCGUCUCCCAGCCCAGCCUCCAAAAUUGCUGUCUUCAU ..........((((...............(((((...((((((...))))))..))))).(((((..(.((((((...((((((........)))))).))))))..)..))))).)))) (-33.42 = -34.02 + 0.60)

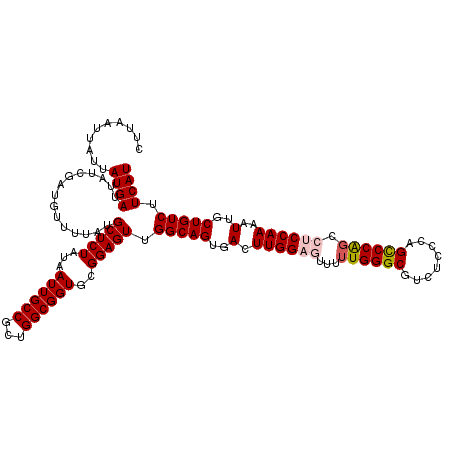
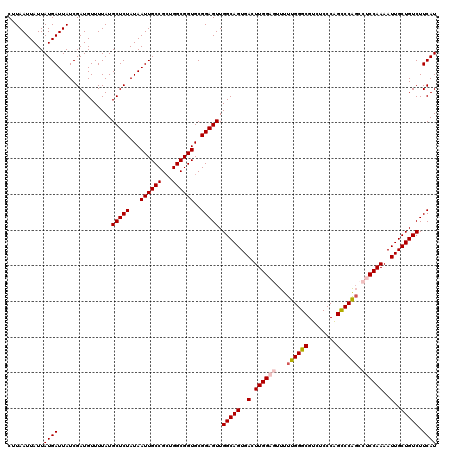
| Location | 17,660,946 – 17,661,066 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.83 |
| Mean single sequence MFE | -38.84 |
| Consensus MFE | -36.62 |
| Energy contribution | -37.06 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.662966 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17660946 120 + 20766785 GCCGCUGGCGGUGCGGAGUUGGCAGUGACUUGGAGUUUUUGGGCGUCUCCCAGCCCAGCCUCCAAAAUUGCUGUCUUCAUUGCGCCUACCUUCUCCUAAGGCCCUUAUCUAAUUGAUAUA (((...)))(((((((.(..(((((..(.((((((...((((((........)))))).))))))..)..)))))..).)))))))..((((.....))))....((((.....)))).. ( -43.10) >DroSec_CAF1 14402 120 + 1 GCCGAUGGCGGUGCGGAGUUGGCAGUGACUUGGAGUUUUUGGGCGUCUCCCAGUCCAGCCUCCAAAAUUGCUGUCUUCAUUGCGCCUACCUUCUCCUAAGGCCCUUACCUAAUUGAUAUA (((...)))(((((((.(..(((((..(.((((((...((((((........)))))).))))))..)..)))))..).)))))))..((((.....))))................... ( -38.90) >DroSim_CAF1 20416 120 + 1 GCCGCUGGCGGUGCGGAGUUGGCAGUGACUUGGAGUUUUUGGGCGUCUCCCAGUCCAGCCUCCAAAAUUGCUGUCUUCAUUGCGCCUACCUUCUCCUAAGGCACUUACCUAAUUGAUAUA (((...((.(((((((.(..(((((..(.((((((...((((((........)))))).))))))..)..)))))..).)))))))........))...))).................. ( -39.20) >DroEre_CAF1 14738 119 + 1 GCCGCUGGCGGUGCGGAGUUGGCAGUGACUUGGACUUUUUGGGCGUCUCCCGGCCCAGCCCCCAAAAUUGCUGUCUUCAUUGCGCCUACCU-UUCCUAAGGCCCUUACCUAAUUGAUAUA (((...)))(((((((.(..(((((..(.((((.....((((((........))))))...))))..)..)))))..).)))))))..(((-(....))))................... ( -36.70) >DroYak_CAF1 26244 120 + 1 GCCGCUGGCGGUGCGGAGUUGGCAGUGACUUGGACUUUUUGGGCGUCUCCCGGCCCGCCCCCCAAAAUUGCUGUCUUCAUUGCGCUUGCCUUUUCCUAAGGCCCUUACCUAAUUGAUAUA (((...)))((((((.(((.(((((..(.((((......(((((........)))))....))))..)..)))))...))).)))..(((((.....)))))....)))........... ( -36.30) >consensus GCCGCUGGCGGUGCGGAGUUGGCAGUGACUUGGAGUUUUUGGGCGUCUCCCAGCCCAGCCUCCAAAAUUGCUGUCUUCAUUGCGCCUACCUUCUCCUAAGGCCCUUACCUAAUUGAUAUA (((...((.((((((.(((.(((((..(.((((((...((((((........)))))).))))))..)..)))))...))).)))..)))....))...))).................. (-36.62 = -37.06 + 0.44)

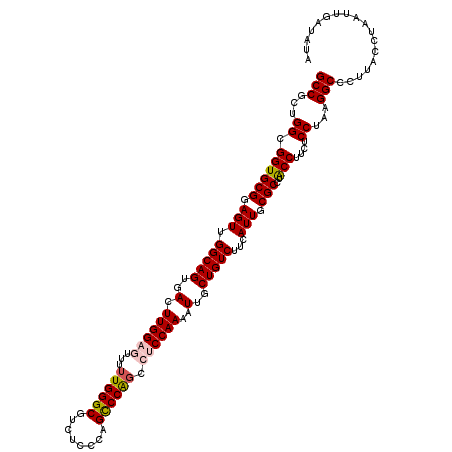
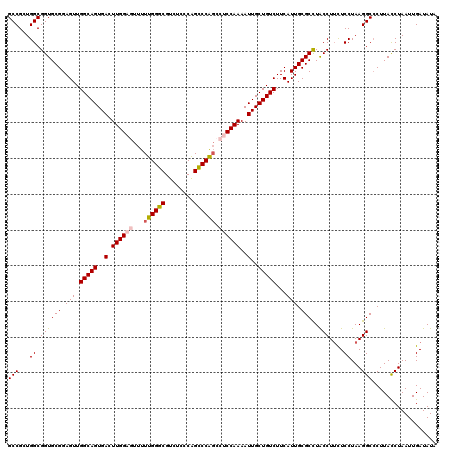
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:47:32 2006