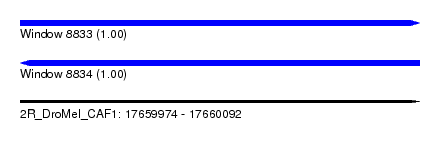
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,659,974 – 17,660,092 |
| Length | 118 |
| Max. P | 0.999599 |

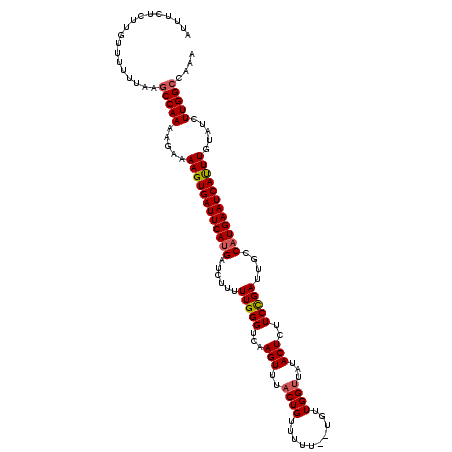
| Location | 17,659,974 – 17,660,092 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.46 |
| Mean single sequence MFE | -26.83 |
| Consensus MFE | -24.28 |
| Energy contribution | -24.56 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.77 |
| SVM RNA-class probability | 0.999599 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17659974 118 + 20766785 AUUUCUCUUGUUUUUUAAGCCAAAGAAAAAGUGAUUCAUGAUCUUUUUGGGUCAAGUUUACUGUUUUU--UGUUGGUUAUACUCUUCCGAUUGCCAUGAAUCAUUUGUAUCUUGGCCAAA ..................(((((.((..((((((((((((......(((((...(((..((((.....--...))))...)))..)))))....))))))))))))...))))))).... ( -28.40) >DroSec_CAF1 13474 119 + 1 AUUUCUCUCGUUUUUUAAACCAAAAGAAAAGUGAUUCAUGAUCUUUUUGGGUCAAGUUUACUGUUUUUUUU-UUGGCUAUACUCUUCCGAUUGCCAUGAAUCAUUUGUAUCUUGGCCAAA ...................((((.....((((((((((((((((....)))))).................-.((((...............)))))))))))))).....))))..... ( -23.56) >DroSim_CAF1 19440 120 + 1 AUUUCUCUUGUUUUUUAAGCCAAAAGAAAAGUGAUUCAUGAUCUUUUUGGGUCAAGUUUACUGUUUUUUUUGUUGGUUAUACUGUUCCGAUUGCCAUGAAUCAUUUGUAUCUUGGCCAAA ..................(((((.....((((((((((((......(((((...(((..((((..........))))...)))..)))))....)))))))))))).....))))).... ( -27.70) >DroEre_CAF1 13774 110 + 1 AAU--------UUCUUAAGCCAAAAGAAAAGUGAUUCAUGAUCUUUUUGGGUCAAGUUUACUGUUUUU--UGUUGGUUAUACUCUUCCGAUUGCCAUGAAUCAUUUGUAUCUUGGCCAGA ...--------.......(((((.....((((((((((((......(((((...(((..((((.....--...))))...)))..)))))....)))))))))))).....))))).... ( -27.50) >DroYak_CAF1 25249 118 + 1 AUUUUUCUUGUUUUUUAUGCCAAAAGAAAAGUGAUUCACGAUCUUUUUGGGUCAAGUUUACUGUUUUU--UGUUGGUUAUACUCUUCUGAUUGCCAUGAAUCACUUGUAUCUUGGCCAAA ..................(((((.....((((((((((.(((((....)))))...............--...((((.((.(......))).)))))))))))))).....))))).... ( -27.00) >consensus AUUUCUCUUGUUUUUUAAGCCAAAAGAAAAGUGAUUCAUGAUCUUUUUGGGUCAAGUUUACUGUUUUU__UGUUGGUUAUACUCUUCCGAUUGCCAUGAAUCAUUUGUAUCUUGGCCAAA ..................(((((.....((((((((((((......(((((...(((..((((..........))))...)))..)))))....)))))))))))).....))))).... (-24.28 = -24.56 + 0.28)

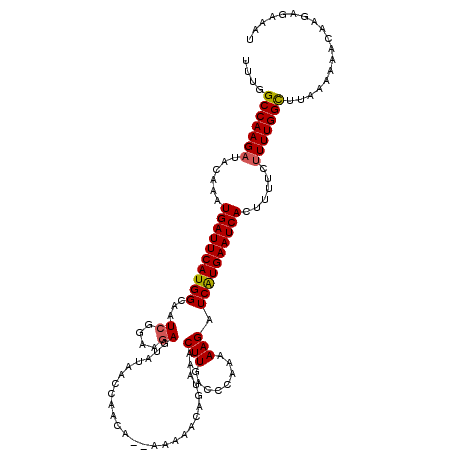
| Location | 17,659,974 – 17,660,092 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.46 |
| Mean single sequence MFE | -23.46 |
| Consensus MFE | -20.20 |
| Energy contribution | -20.28 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.71 |
| SVM RNA-class probability | 0.999547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17659974 118 - 20766785 UUUGGCCAAGAUACAAAUGAUUCAUGGCAAUCGGAAGAGUAUAACCAACA--AAAAACAGUAAACUUGACCCAAAAAGAUCAUGAAUCACUUUUUCUUUGGCUUAAAAAACAAGAGAAAU ...((((((((...((.((((((((((...((....))............--............(((........))).)))))))))).))..)).))))))................. ( -22.90) >DroSec_CAF1 13474 119 - 1 UUUGGCCAAGAUACAAAUGAUUCAUGGCAAUCGGAAGAGUAUAGCCAA-AAAAAAAACAGUAAACUUGACCCAAAAAGAUCAUGAAUCACUUUUCUUUUGGUUUAAAAAACGAGAGAAAU .................(((((((((((..((....)).....)))..-...............(((........)))...))))))))..(((((((((..........))))))))). ( -22.90) >DroSim_CAF1 19440 120 - 1 UUUGGCCAAGAUACAAAUGAUUCAUGGCAAUCGGAACAGUAUAACCAACAAAAAAAACAGUAAACUUGACCCAAAAAGAUCAUGAAUCACUUUUCUUUUGGCUUAAAAAACAAGAGAAAU ...((((((((......((((((((((.....((..........))..................(((........))).))))))))))......))))))))................. ( -21.10) >DroEre_CAF1 13774 110 - 1 UCUGGCCAAGAUACAAAUGAUUCAUGGCAAUCGGAAGAGUAUAACCAACA--AAAAACAGUAAACUUGACCCAAAAAGAUCAUGAAUCACUUUUCUUUUGGCUUAAGAA--------AUU (((((((((((......((((((((((...((....))............--............(((........))).))))))))))......))))))))..))).--------... ( -25.00) >DroYak_CAF1 25249 118 - 1 UUUGGCCAAGAUACAAGUGAUUCAUGGCAAUCAGAAGAGUAUAACCAACA--AAAAACAGUAAACUUGACCCAAAAAGAUCGUGAAUCACUUUUCUUUUGGCAUAAAAAACAAGAAAAAU ....(((((((.(.(((((((((((((...((....))............--............(((........))).))))))))))))).).))))))).................. ( -25.40) >consensus UUUGGCCAAGAUACAAAUGAUUCAUGGCAAUCGGAAGAGUAUAACCAACA__AAAAACAGUAAACUUGACCCAAAAAGAUCAUGAAUCACUUUUCUUUUGGCUUAAAAAACAAGAGAAAU ....(((((((......((((((((((...((....))..........................(((........))).))))))))))......))))))).................. (-20.20 = -20.28 + 0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:47:24 2006