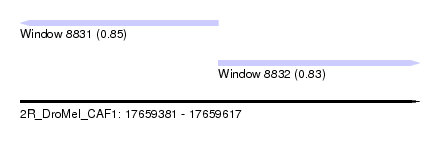
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,659,381 – 17,659,617 |
| Length | 236 |
| Max. P | 0.854861 |

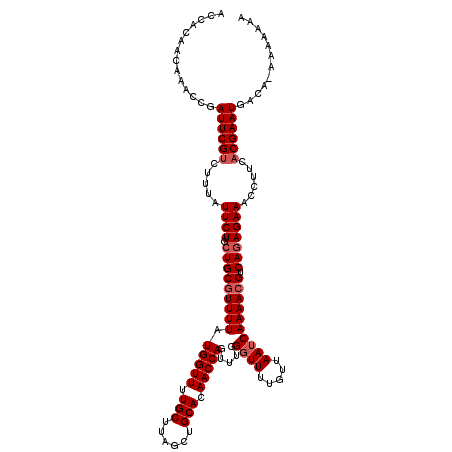
| Location | 17,659,381 – 17,659,498 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.60 |
| Mean single sequence MFE | -32.08 |
| Consensus MFE | -25.44 |
| Energy contribution | -24.20 |
| Covariance contribution | -1.24 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.854861 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17659381 117 - 20766785 GGAAAGGUGCCCGAAGAUCUCCAACAAAACGCUCAUAACUGGAGCAUGAAUGCUUAUUUGAGCGGGGAUUUUAUGAUGAUUU-UUU--CAGUUCCUUUGGCAGCCUAUCAUCUAUCGACC .((.(((((((..(((((((((........((((.......))))......((((....)))))))))))))....(((...-..)--))........)))).))).))........... ( -31.80) >DroSec_CAF1 12889 115 - 1 GGAAAGGUGCCCAAAGAUCUCCAACAAAACGCUCAUAACUGGAGCAUGAAUGCUUAUUUAAGCGGGGAUUUUAUGAUGAUUUUUUU--CAGUUCUUUUGGCAGCCUU---UCUAUCGACC (((((((((((.((((((((((........((((.......))))......((((....)))))))))........(((......)--))..))))).)))).))))---)))....... ( -36.30) >DroSim_CAF1 18854 114 - 1 GGAAAGGUGCCCAAAGAUCUCCAACAAAACGCUCAUAACUGGAGCAUGAAUGCUUAUUUAAGCGGGGAUUUUAUGAUGAUUU-UUU--CAGAUCUUUUGGCAGCCUA---UCUAUUGACC (((.(((((((.((((((((((........((((.......)))).....(((((....))))).)))........(((...-..)--))))))))).)))).))).---)))....... ( -33.40) >DroEre_CAF1 13187 114 - 1 GGAAAGGUGGCCGAAGAUCUUUAACAAAACGCUCAUAACUGGAGCAUGAAUGCUUAUUUAACCGGGGAUUUUAUGAUUAUUU-UUU--CAGUAUUUUUGGGAGCCUG---UCCAUCGACG (((.((((..((((((((((..........((((.......))))..((((.(((........))).))))...........-...--.)).))))))))..)))).---)))....... ( -25.60) >DroYak_CAF1 24660 115 - 1 GGAAAGGUGGCCGAAGAUCCUCAA-AUAACGCUCAUAACUGGAGCAUGAAUGCUUAUUUAAGCGGGGAUUUUAUGAUAAUUU-UUUUGUAGUUCUUUUGGGAGCCUG---UCCAUCGACC (((.((((..((((((((((((..-.....((((.......))))......((((....)))))))))))(((..(......-..)..)))....)))))..)))).---)))....... ( -33.30) >consensus GGAAAGGUGCCCGAAGAUCUCCAACAAAACGCUCAUAACUGGAGCAUGAAUGCUUAUUUAAGCGGGGAUUUUAUGAUGAUUU_UUU__CAGUUCUUUUGGCAGCCUA___UCUAUCGACC (((.((((..((((((((((((........((((.......))))......((((....))))))))))))).....((((........))))....)))..))))....)))....... (-25.44 = -24.20 + -1.24)

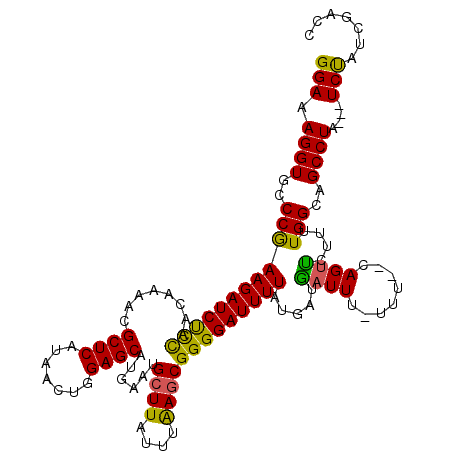
| Location | 17,659,498 – 17,659,617 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.65 |
| Mean single sequence MFE | -25.90 |
| Consensus MFE | -22.76 |
| Energy contribution | -23.56 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.832151 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17659498 119 + 20766785 ACCACAACAAACCGAUUCGUCUUUAUUCUGCUGCAUUUUAUGGUUUUGCUUAGCUGCACAACCAGUUUGGGUUUUGUUAAUCAAAACGUCAGAGAAACCUUCACGAAUGACA-AAAAAAA ..............((((((.....((((.((((......(((((.(((......))).)))))......((((((.....))))))).)))))))......))))))....-....... ( -24.40) >DroSec_CAF1 13004 118 + 1 ACCACAACAAACCGAUUCGUCUUUAUUCUGGUGCGUUUUAUGGUUUUGCUUAGCUGCACAACCAGUUUGGGUUUUGUUAAUCAAAACGUCAGAGAAACCUUCACGAAUGACA-AAA-AAA ..................(((...(((((((.(.(((((.(((((.(((......))).))))).(((((((((((.....)))))).))))))))))).))).))))))).-...-... ( -25.50) >DroSim_CAF1 18968 118 + 1 ACCACAACAAACCGAUUCGUCUUUAUUCUGCUGCGUUUUAUGGUUUUGCUUAGCUGCACAACCAGUUUGGGUUUUGUUAAUCAAAACGUCAGAGAAACCUUCACGAAUGACA-AAA-AAA ..............((((((.....((((.(((((((((.(((((.(((......))).))))).....((((.....)))))))))).)))))))......))))))....-...-... ( -26.30) >DroEre_CAF1 13301 119 + 1 ACCACAACAAACCGAUUCGUUUUUAUUCUGCUGCGUUUUAUGGUUUUGCUUAGCUGCACAACCAGUUUGGGUUUUGUUAAUCAAAACGUCAGAGAAACCUUUUCGAAUGACA-GAAAAAA .(((.(((.....((..(((............)))..)).(((((.(((......))).)))))))))))((((((.....))))))((((..(((.....)))...)))).-....... ( -25.50) >DroYak_CAF1 24775 120 + 1 ACCACAACAAACCGAUUCGUCUUUAUUCUGCUGCGUUUUAUGGUUUUGCUUAGCUGCACAACCAGUUUGGCUUUUGUUAAUCAAAACGUCAGAGAAACCUUCACGAAUGACAAGAAAAAA ..............((((((.....((((.(((((((((.(((((.(((......))).)))))(.(((((....))))).))))))).)))))))......))))))............ ( -27.80) >consensus ACCACAACAAACCGAUUCGUCUUUAUUCUGCUGCGUUUUAUGGUUUUGCUUAGCUGCACAACCAGUUUGGGUUUUGUUAAUCAAAACGUCAGAGAAACCUUCACGAAUGACA_AAAAAAA ..............((((((.....((((.(((((((((.(((((.(((......))).))))).....((((.....)))))))))).)))))))......))))))............ (-22.76 = -23.56 + 0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:47:22 2006