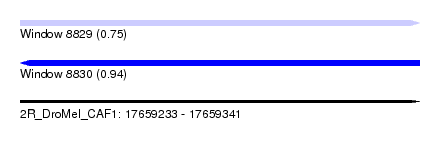
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,659,233 – 17,659,341 |
| Length | 108 |
| Max. P | 0.940489 |

| Location | 17,659,233 – 17,659,341 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.00 |
| Mean single sequence MFE | -26.05 |
| Consensus MFE | -23.58 |
| Energy contribution | -23.10 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.748790 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17659233 108 + 20766785 AGAUUCUGGGCAGCUCACUUCUAAG------------ACUGGGAACCCAUUAAGUGGUAAGUGCUAAAAACGUAAAAAUAUUUAACAACCCGUGGUAUUUGUCAAACAUUUAAGCUGCCA ........(((((((..........------------..(((....))).....((..((((((((......((((....))))........))))))))..))........))))))). ( -25.34) >DroSec_CAF1 12741 108 + 1 AGAUUCUGGGCAGCUCACUUCUAAG------------ACUGGAAACCCAUUAAGUGGUAAGUGCUAAAAACGUAAAAAAAUUUAACAACCCGUGGCAUUUGUCAAACAUUUAAGCUGCCA ........(((((((...(((((..------------..)))))..........((..((((((((......((((....))))........))))))))..))........))))))). ( -26.14) >DroSim_CAF1 18706 108 + 1 AGAUUCUGGGCAGCUCACUUCUAAG------------ACUGGAAACCCAUUAAGUGGUAAGUGCUAAAAACGUAAAAAUAUUUAACAACCCGUGGUAUUUGUCAAACAUUUAAGCUGCCA ........(((((((...(((((..------------..)))))..........((..((((((((......((((....))))........))))))))..))........))))))). ( -24.14) >DroEre_CAF1 13027 120 + 1 AGAUCCUGGGCAGCUCACUUCUAACACUGGAGUCGUCACUGGAAACCCAUUAAGUGGUAAGUGCUAGGAACGUAAAAAUAUUUAACAACCCGUGGUGUUUGUCAAACACAUAAGCUGCCA ........(((((((.........(((.((...(((..((((....((((...))))......))))..)))((((....))))....)).)))(((((.....)))))...))))))). ( -30.50) >DroYak_CAF1 24500 120 + 1 AGAUUCUGGGCAGUUCACUUCUAACGCUGGAGUCAUCACUGGAAACCCAUUAACUGGUAAGUGCUAAAAACGUAAAAAUAUUUAACAGCCCGUGGUAUUUGUCAAACAUUUAAGCUGCCA ........(((((((.(((((.......)))))......(((....))).....((..((((((((......((((....))))........))))))))..))........))))))). ( -24.14) >consensus AGAUUCUGGGCAGCUCACUUCUAAG____________ACUGGAAACCCAUUAAGUGGUAAGUGCUAAAAACGUAAAAAUAUUUAACAACCCGUGGUAUUUGUCAAACAUUUAAGCUGCCA ........(((((((........................(((....))).....((..((((((((......((((....))))........))))))))..))........))))))). (-23.58 = -23.10 + -0.48)

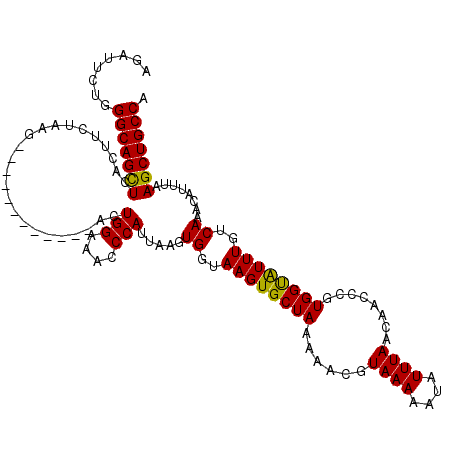
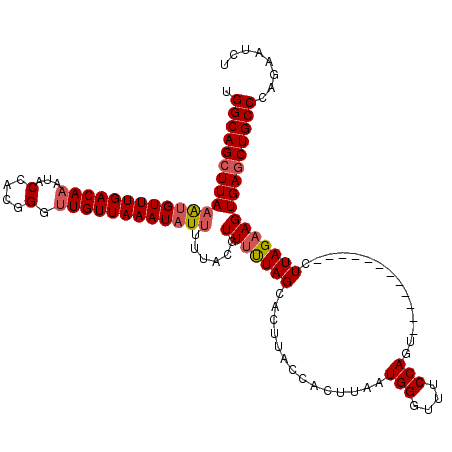
| Location | 17,659,233 – 17,659,341 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.00 |
| Mean single sequence MFE | -30.94 |
| Consensus MFE | -24.35 |
| Energy contribution | -25.03 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940489 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17659233 108 - 20766785 UGGCAGCUUAAAUGUUUGACAAAUACCACGGGUUGUUAAAUAUUUUUACGUUUUUAGCACUUACCACUUAAUGGGUUCCCAGU------------CUUAGAAGUGAGCUGCCCAGAAUCU .(((((((.(((((((((((((...(....).)))))))))))))............(((((.........(((....)))..------------.....))))))))))))........ ( -31.19) >DroSec_CAF1 12741 108 - 1 UGGCAGCUUAAAUGUUUGACAAAUGCCACGGGUUGUUAAAUUUUUUUACGUUUUUAGCACUUACCACUUAAUGGGUUUCCAGU------------CUUAGAAGUGAGCUGCCCAGAAUCU .(((((((((..(.(..(((..........((((((((((............)))))))...)))......(((....)))))------------)..).)..)))))))))........ ( -27.30) >DroSim_CAF1 18706 108 - 1 UGGCAGCUUAAAUGUUUGACAAAUACCACGGGUUGUUAAAUAUUUUUACGUUUUUAGCACUUACCACUUAAUGGGUUUCCAGU------------CUUAGAAGUGAGCUGCCCAGAAUCU .(((((((((((((((((((((...(....).))))))))))))......(((((((.(((.(((.(.....))))....)))------------.))))))))))))))))........ ( -30.70) >DroEre_CAF1 13027 120 - 1 UGGCAGCUUAUGUGUUUGACAAACACCACGGGUUGUUAAAUAUUUUUACGUUCCUAGCACUUACCACUUAAUGGGUUUCCAGUGACGACUCCAGUGUUAGAAGUGAGCUGCCCAGGAUCU .(((((((((((((((((((((...(....).)))))))))))..........((((((((..(((.....)))(((......)))......))))))))..))))))))))........ ( -37.70) >DroYak_CAF1 24500 120 - 1 UGGCAGCUUAAAUGUUUGACAAAUACCACGGGCUGUUAAAUAUUUUUACGUUUUUAGCACUUACCAGUUAAUGGGUUUCCAGUGAUGACUCCAGCGUUAGAAGUGAACUGCCCAGAAUCU .(((((...((((((((((((....(....)..))))))))))))...((((((((((.((....(((((.(((....)))....)))))..)).))))))))))..)))))........ ( -27.80) >consensus UGGCAGCUUAAAUGUUUGACAAAUACCACGGGUUGUUAAAUAUUUUUACGUUUUUAGCACUUACCACUUAAUGGGUUUCCAGU____________CUUAGAAGUGAGCUGCCCAGAAUCU .(((((((((((((((((((((...(....).))))))))))))......(((((((..............(((....)))...............))))))))))))))))........ (-24.35 = -25.03 + 0.68)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:47:20 2006