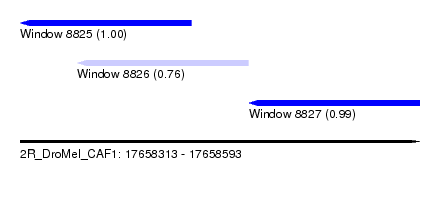
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,658,313 – 17,658,593 |
| Length | 280 |
| Max. P | 0.997573 |

| Location | 17,658,313 – 17,658,433 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.17 |
| Mean single sequence MFE | -24.64 |
| Consensus MFE | -22.06 |
| Energy contribution | -22.06 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.89 |
| SVM RNA-class probability | 0.997573 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17658313 120 - 20766785 CUACAGUUCUUUUUGAACAGUUCUUAAAUCCAACCGGUUCUGAUAUGAUUAUUAUUAUGCUCCAACCGGUUUGCAAGGCAAACAUUUUAAUUUUAUUCGGUUUUAUUUUGCAUUGUUCCU ..............(((((((.(.((((.((...(((((..((((((((....)))))).)).)))))((((((...))))))...............)).))))....).))))))).. ( -23.10) >DroSec_CAF1 11849 119 - 1 CUACAGGUCUUUUUGAACAGUUCUUAAAUCCAACCGGUUCUCAUAUGAUUAUUAUUAUGCUCGAACCGGUUUGCAAGGCAAAC-UUUUAAUUUUAUUCGGUUUUAUUUUGCAUUGUUCCU ..............(((((((.(..((((..(((((((((...((((((....))))))...)))))(((((((...))))))-).............))))..)))).).))))))).. ( -26.10) >DroSim_CAF1 17801 120 - 1 CUACAGUUCUUUUUGAACAGUUCUUAAAUCCAACCGGUUCUGAUAUGAUUAUUAUUAUGCUCGAACCGGUUUGCAAGGCAAACAUUUUAAUUUUAUUCGGUUUUAUUUUGCAUUGUUCCU ..............(((((((.(.((((.((...((((((.((((((((....)))))).))))))))((((((...))))))...............)).))))....).))))))).. ( -27.10) >DroEre_CAF1 12111 120 - 1 CUACAGUUCUUUUUGAACAGUUCUUAAAUCCAACCGGUUCUGAUAUGAUUAUUAUUAUGCCUCAACCGGUUUGCAAGGCAAACAUUUUAAUUUUAUUCGGUUUUAUUUUGCAUUGUUCCU ..............(((((((.(.((((.((...(((((..(.((((((....)))))).)..)))))((((((...))))))...............)).))))....).))))))).. ( -23.80) >DroYak_CAF1 23551 120 - 1 CUACAGUUCAUUUUGAACAGUUCUUAAAUCCAACCGGUUCUGAUAUGAUUAUUAUUAUGCUCCAACCGGUUUGCAAGGCAAACAUUUUAAUUUUAUUCGGUUUUAUUUUGCAUUGUUCCC ..............(((((((.(.((((.((...(((((..((((((((....)))))).)).)))))((((((...))))))...............)).))))....).))))))).. ( -23.10) >consensus CUACAGUUCUUUUUGAACAGUUCUUAAAUCCAACCGGUUCUGAUAUGAUUAUUAUUAUGCUCCAACCGGUUUGCAAGGCAAACAUUUUAAUUUUAUUCGGUUUUAUUUUGCAUUGUUCCU ..............(((((((.(.((((.((...(((((....((((((....))))))....)))))((((((...))))))...............)).))))....).))))))).. (-22.06 = -22.06 + 0.00)



| Location | 17,658,353 – 17,658,473 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.58 |
| Mean single sequence MFE | -31.12 |
| Consensus MFE | -24.54 |
| Energy contribution | -25.34 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.760273 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17658353 120 - 20766785 CUAAGCCAGAAAAACUGUGGCGACUUUAUUCUGGGUCUGACUACAGUUCUUUUUGAACAGUUCUUAAAUCCAACCGGUUCUGAUAUGAUUAUUAUUAUGCUCCAACCGGUUUGCAAGGCA ....((((((((((((((((((((((......))))).).)))))))).))))).................((((((((..((((((((....)))))).)).)))))))).....))). ( -33.40) >DroSec_CAF1 11888 120 - 1 CUAAGGCAGAAAAGCUGUGGCGACUUUAUUCUGGGUCUGACUACAGGUCUUUUUGAACAGUUCUUAAAUCCAACCGGUUCUCAUAUGAUUAUUAUUAUGCUCGAACCGGUUUGCAAGGCA .....(((((((((((((((((((((......))))).).))))))..)))))).................(((((((((...((((((....))))))...))))))))))))...... ( -32.70) >DroSim_CAF1 17841 120 - 1 CUAAGGCUGAAAAGCUGUGGCGACUUUAUUCUGGGUCUGACUACAGUUCUUUUUGAACAGUUCUUAAAUCCAACCGGUUCUGAUAUGAUUAUUAUUAUGCUCGAACCGGUUUGCAAGGCA ....((((....))))((.((((........(((((.(((..((.((((.....)))).))..))).)))))((((((((.((((((((....)))))).))))))))))))))...)). ( -35.60) >DroEre_CAF1 12151 120 - 1 CAAAGGCAGAAAAACUGUGGCGAUUUUAUUCUGGCUCUGACUACAGUUCUUUUUGAACAGUUCUUAAAUCCAACCGGUUCUGAUAUGAUUAUUAUUAUGCCUCAACCGGUUUGCAAGGCA .....((((.....)))).((((........(((...(((..((.((((.....)))).))..)))...)))(((((((..(.((((((....)))))).)..)))))))))))...... ( -25.70) >DroYak_CAF1 23591 120 - 1 CCAAGGCAGAAAAACUGUGGAGAUUUUAUUCUCUGUUCGACUACAGUUCAUUUUGAACAGUUCUUAAAUCCAACCGGUUCUGAUAUGAUUAUUAUUAUGCUCCAACCGGUUUGCAAGGCA ((...(((((.......((((...((((....((((((((............))))))))....)))))))).((((((..((((((((....)))))).)).)))))))))))..)).. ( -28.20) >consensus CUAAGGCAGAAAAACUGUGGCGACUUUAUUCUGGGUCUGACUACAGUUCUUUUUGAACAGUUCUUAAAUCCAACCGGUUCUGAUAUGAUUAUUAUUAUGCUCCAACCGGUUUGCAAGGCA ......((((((((((((((.(((((......)))))...)))))))).))))))....((.(((....(.((((((((....((((((....))))))....)))))))).).))))). (-24.54 = -25.34 + 0.80)

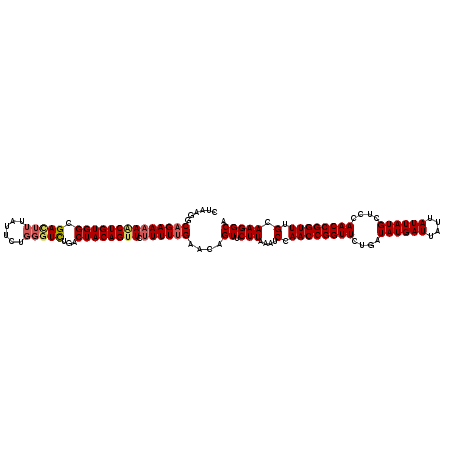

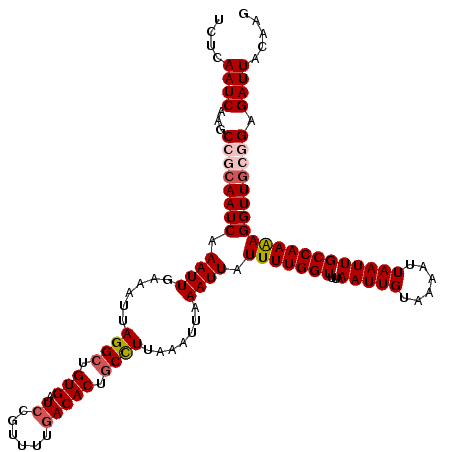
| Location | 17,658,473 – 17,658,593 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -27.48 |
| Consensus MFE | -24.22 |
| Energy contribution | -25.30 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.994392 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17658473 120 - 20766785 UCUCAAUCAAGCCGCAAUCAAAUUGAAAUUAGGCUGUGAUCCGUUGGAACACUGCUUUAAAUUAAAUUAUUUUGGUUUAAAUUGUAAAAUUAAUUGCCAAAAGGUUGCGGAGAUUACAAG ....((((...((((((((...((((....((((.(((.(((...))).))).))))....))))....(((((((...(((((......)))))))))))))))))))).))))..... ( -31.90) >DroSec_CAF1 12008 120 - 1 UCUCAAUCAAGCCGCAAUCAAAUUGAAAUUAGGCUGUGAUCCGUUUGGACACUGCCUUAAAGUGAAUUAUUUUGGUUUAAAUUGUAAAAUUAAUUGCCAAAAGGUUGCAGAGAUUACAAG ....((((...(.((((((.((((.(....((((.(((.(((....)))))).)))).....).)))).(((((((...(((((......)))))))))))))))))).).))))..... ( -29.70) >DroSim_CAF1 17961 120 - 1 UCUCAAUCAAGCCGCAAUCAAAUUGAAAUUAGGCUGUGAUCCUUUUUGACACUGCCUUAAAUUAAAUUAUUUUGGUUUAAAUUGUAAAAUUAAUUGCCAAAAGGUUGCGGAGAUUACAAG ....((((...((((((((...((((....((((.(((.((......))))).))))....))))....(((((((...(((((......)))))))))))))))))))).))))..... ( -31.90) >DroEre_CAF1 12271 120 - 1 UCUCAAUCAAGCCCCAAUCAAAUUGAAAUUAGGCUGUGAUCCGUUUUGACACUGCCUUAAAUUAAAUUAUUUUGGUUUAAAUUGUAAAAUUAAUUGCCAAGAGGUUGCGGAGAUUACAAG ....((((...((.(((((...((((....((((.(((.((......))))).))))....))))....(((((((...(((((......))))))))))))))))).)).))))..... ( -25.70) >DroYak_CAF1 23711 120 - 1 UCUCAAUCAAGCGCCAAUCAAAUUGAAAUUAGGCUGUGAUCCGUUGUGACACUACCUUAAAUUAAAUUAUUUUGGUUUAAAUUGUAAAAUUAAUUGCCAAAAGGUUUCAGAGAUUACAAG ...............((((...(((((((((((..(((.((......)))))..)))............(((((((...(((((......)))))))))))))))))))).))))..... ( -18.20) >consensus UCUCAAUCAAGCCGCAAUCAAAUUGAAAUUAGGCUGUGAUCCGUUUUGACACUGCCUUAAAUUAAAUUAUUUUGGUUUAAAUUGUAAAAUUAAUUGCCAAAAGGUUGCGGAGAUUACAAG ....((((...((((((((.((((......((((.(((.((......))))).)))).......)))).(((((((...(((((......)))))))))))))))))))).))))..... (-24.22 = -25.30 + 1.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:47:17 2006