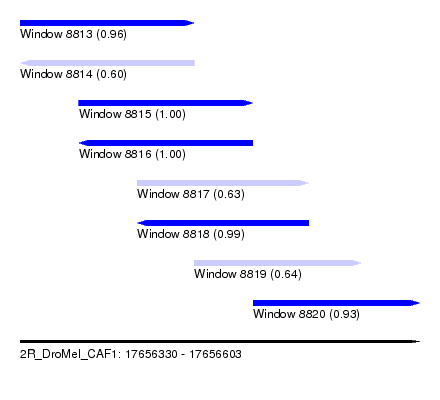
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,656,330 – 17,656,603 |
| Length | 273 |
| Max. P | 0.999932 |

| Location | 17,656,330 – 17,656,449 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.31 |
| Mean single sequence MFE | -38.34 |
| Consensus MFE | -36.08 |
| Energy contribution | -35.72 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958847 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17656330 119 + 20766785 CUAAUUGGAGUUGGCCAAGUCGGAAAAUGCGGUAACUGCAGCGCAGGCGGGAUUCUCGCCGAGAAACGAGAAAUCUUCGCAUCUCCU-UGCGGAGGAAAGCGAAAGUUAGACGCUUUAAA ....((((......))))..........(((.(((((....(((.((((((...)))))).............((((((((......-))))))))...)))..)))))..)))...... ( -35.80) >DroSec_CAF1 9873 119 + 1 CUAAUUGGAGUUGGCCAAGUCGGAAAAUGCGGUAACUGCAGCGCUGGCGGGAUUCUCGGCGAGAAACGAGAAAUCUCCGCAUCUCCU-UGCGGAGGAAAGCGAAAGUUAGACGCUUUAAA ....((((......))))(((((....(((((...)))))...)))))....((((((........)))))).((((((((......-))))))))((((((.........))))))... ( -40.40) >DroSim_CAF1 15826 119 + 1 CUAAUUGGAGUUGGCCAAGUCGGAAAAUGCGGUAACUGCAGCGCUGGCGGGAUUCUCGGCGAGAAACGAGAAAUCUCCGCAUCUCCU-UGCGGAGGAAAGCGAAAGUUAGACGCUUUAAA ....((((......))))(((((....(((((...)))))...)))))....((((((........)))))).((((((((......-))))))))((((((.........))))))... ( -40.40) >DroEre_CAF1 10114 119 + 1 CUAAUUGGAGUUGGCCAAGUCGGAAAAUGCGGUAACUGCAAUGCAGGCGGGAUUCUCGGCGAGAAACGUGCAAUCUCCGCAUCUCCU-UGCGGAGGAAAGCGAAAGUUAGACGCUUUAAA ((.((((.(((((.((..((......))..))))))).))))..))(((.(.(((((...))))).).)))..((((((((......-))))))))((((((.........))))))... ( -36.50) >DroYak_CAF1 21565 120 + 1 CUAAUUGGAGUUGGCCAAGUCGGAAAAUGCGGUAACUGCCGUGCAGGCGGGAUUCUUGGCGAGAAACGAGAAAUCUCCGCAUCUCCUUUGCGGAGGAAAGCGAAAGUUAGACGCUUUAAA ....(((...((.(((((((((.......)))...(((((.....)))))....)))))).))...)))....((((((((.......))))))))((((((.........))))))... ( -38.60) >consensus CUAAUUGGAGUUGGCCAAGUCGGAAAAUGCGGUAACUGCAGCGCAGGCGGGAUUCUCGGCGAGAAACGAGAAAUCUCCGCAUCUCCU_UGCGGAGGAAAGCGAAAGUUAGACGCUUUAAA ....((((......))))..........(((.(((((....(((..((((((((((((........)))).)))).)))).(((((.....)))))...)))..)))))..)))...... (-36.08 = -35.72 + -0.36)

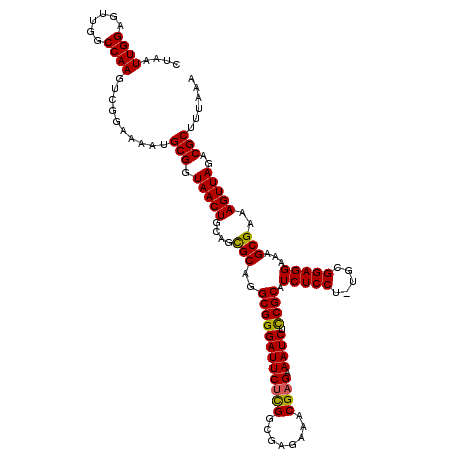
| Location | 17,656,330 – 17,656,449 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.31 |
| Mean single sequence MFE | -33.62 |
| Consensus MFE | -27.26 |
| Energy contribution | -28.26 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.602666 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17656330 119 - 20766785 UUUAAAGCGUCUAACUUUCGCUUUCCUCCGCA-AGGAGAUGCGAAGAUUUCUCGUUUCUCGGCGAGAAUCCCGCCUGCGCUGCAGUUACCGCAUUUUCCGACUUGGCCAACUCCAAUUAG ......(((..(((((..(((....((((...-.))))..(((..((((.(((((......))))))))).)))..)))....))))).)))..........((((......)))).... ( -32.50) >DroSec_CAF1 9873 119 - 1 UUUAAAGCGUCUAACUUUCGCUUUCCUCCGCA-AGGAGAUGCGGAGAUUUCUCGUUUCUCGCCGAGAAUCCCGCCAGCGCUGCAGUUACCGCAUUUUCCGACUUGGCCAACUCCAAUUAG ......(((..(((((..((((...((((...-.))))..((((.((((.((((........)))))))))))).))))....))))).)))..........((((......)))).... ( -36.10) >DroSim_CAF1 15826 119 - 1 UUUAAAGCGUCUAACUUUCGCUUUCCUCCGCA-AGGAGAUGCGGAGAUUUCUCGUUUCUCGCCGAGAAUCCCGCCAGCGCUGCAGUUACCGCAUUUUCCGACUUGGCCAACUCCAAUUAG ......(((..(((((..((((...((((...-.))))..((((.((((.((((........)))))))))))).))))....))))).)))..........((((......)))).... ( -36.10) >DroEre_CAF1 10114 119 - 1 UUUAAAGCGUCUAACUUUCGCUUUCCUCCGCA-AGGAGAUGCGGAGAUUGCACGUUUCUCGCCGAGAAUCCCGCCUGCAUUGCAGUUACCGCAUUUUCCGACUUGGCCAACUCCAAUUAG ...((((((.........)))))).((((...-.))))((((((.(((((((...(((((...)))))....(....)..))))))).))))))........((((......)))).... ( -33.90) >DroYak_CAF1 21565 120 - 1 UUUAAAGCGUCUAACUUUCGCUUUCCUCCGCAAAGGAGAUGCGGAGAUUUCUCGUUUCUCGCCAAGAAUCCCGCCUGCACGGCAGUUACCGCAUUUUCCGACUUGGCCAACUCCAAUUAG ....(((((...((.((((((((((((......)))))).)))))).))...)))))...((((((......(((.....))).((....)).........))))))............. ( -29.50) >consensus UUUAAAGCGUCUAACUUUCGCUUUCCUCCGCA_AGGAGAUGCGGAGAUUUCUCGUUUCUCGCCGAGAAUCCCGCCUGCGCUGCAGUUACCGCAUUUUCCGACUUGGCCAACUCCAAUUAG ......(((..(((((..(((....((((.....))))..((((.((((.((((........))))))))))))..)))....))))).)))..........((((......)))).... (-27.26 = -28.26 + 1.00)

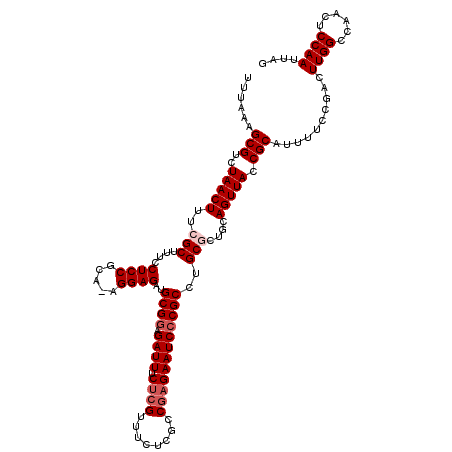
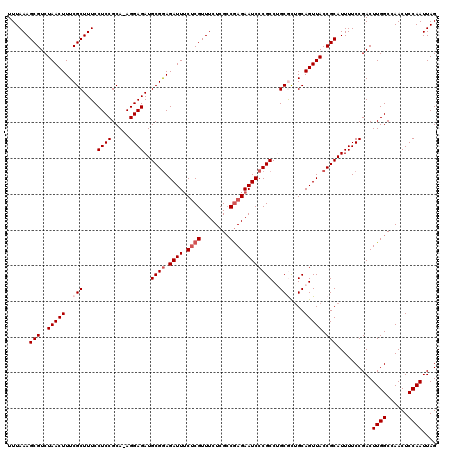
| Location | 17,656,370 – 17,656,489 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.64 |
| Mean single sequence MFE | -42.00 |
| Consensus MFE | -38.90 |
| Energy contribution | -39.18 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.93 |
| SVM decision value | 4.64 |
| SVM RNA-class probability | 0.999932 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17656370 119 + 20766785 GCGCAGGCGGGAUUCUCGCCGAGAAACGAGAAAUCUUCGCAUCUCCU-UGCGGAGGAAAGCGAAAGUUAGACGCUUUAAAUUUGAAAUUGAAAUUUCUUGUUUCUCGGUUUAUGGCCGAG .....((((((...))))))(((((((((((((((((((((......-))))))))((((((.........))))))................)))))))))))))(((.....)))... ( -45.00) >DroSec_CAF1 9913 119 + 1 GCGCUGGCGGGAUUCUCGGCGAGAAACGAGAAAUCUCCGCAUCUCCU-UGCGGAGGAAAGCGAAAGUUAGACGCUUUAAAUUUGAAAUUGAAAUUUCUUGUUUCUCGGUUUAUGGCCGAG ((....))......(((((((((((((((((((((((((((......-))))))))((((((.........))))))................)))))))))))))........)))))) ( -44.30) >DroSim_CAF1 15866 119 + 1 GCGCUGGCGGGAUUCUCGGCGAGAAACGAGAAAUCUCCGCAUCUCCU-UGCGGAGGAAAGCGAAAGUUAGACGCUUUAAAUUUGAAAUUGAAAUUUCUUGUUUCUCGGUUUAUGGCCGAG ((....))......(((((((((((((((((((((((((((......-))))))))((((((.........))))))................)))))))))))))........)))))) ( -44.30) >DroEre_CAF1 10154 119 + 1 AUGCAGGCGGGAUUCUCGGCGAGAAACGUGCAAUCUCCGCAUCUCCU-UGCGGAGGAAAGCGAAAGUUAGACGCUUUAAAUUUAAAAUUGAAAUUUCUUGUUUCUCGGCUUAUGGCCGAG .......((((...))))((((((((....(((((((((((......-))))))))((((((.........))))))..........)))...))))))))..((((((.....)))))) ( -38.20) >DroYak_CAF1 21605 120 + 1 GUGCAGGCGGGAUUCUUGGCGAGAAACGAGAAAUCUCCGCAUCUCCUUUGCGGAGGAAAGCGAAAGUUAGACGCUUUAAAUUUGAAAUUGAAAUUUCUUGUUACUCGGUUUAUGGCCGAG .....(((.(.......(.((((.(((((((((((((((((.......))))))))((((((.........))))))................))))))))).)))).)...).)))... ( -38.20) >consensus GCGCAGGCGGGAUUCUCGGCGAGAAACGAGAAAUCUCCGCAUCUCCU_UGCGGAGGAAAGCGAAAGUUAGACGCUUUAAAUUUGAAAUUGAAAUUUCUUGUUUCUCGGUUUAUGGCCGAG .....((((((...)))..((((((((((((((((((((((.......))))))))((((((.........))))))................)))))))))))))).......)))... (-38.90 = -39.18 + 0.28)

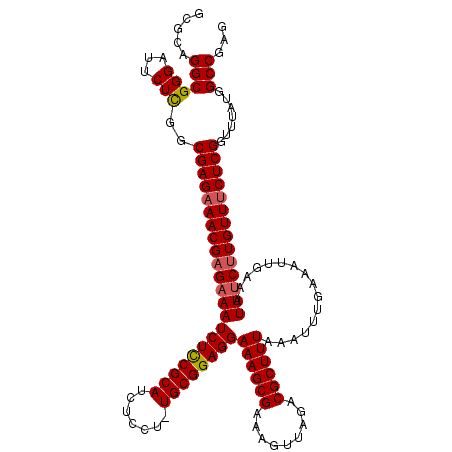
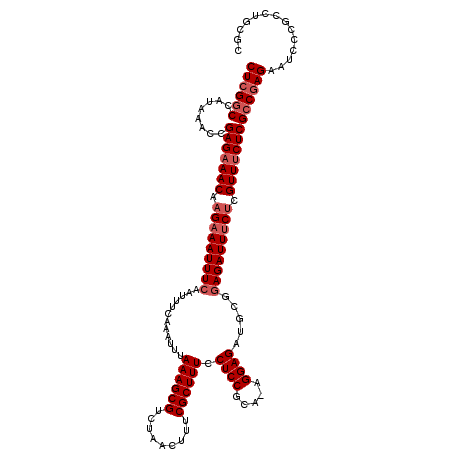
| Location | 17,656,370 – 17,656,489 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.64 |
| Mean single sequence MFE | -37.60 |
| Consensus MFE | -32.84 |
| Energy contribution | -34.04 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.69 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.97 |
| SVM RNA-class probability | 0.999735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17656370 119 - 20766785 CUCGGCCAUAAACCGAGAAACAAGAAAUUUCAAUUUCAAAUUUAAAGCGUCUAACUUUCGCUUUCCUCCGCA-AGGAGAUGCGAAGAUUUCUCGUUUCUCGGCGAGAAUCCCGCCUGCGC ....((........(((((((.(((((((((............((((((.........)))))).((((...-.))))....).)))))))).)))))))((((.......)))).)).. ( -38.00) >DroSec_CAF1 9913 119 - 1 CUCGGCCAUAAACCGAGAAACAAGAAAUUUCAAUUUCAAAUUUAAAGCGUCUAACUUUCGCUUUCCUCCGCA-AGGAGAUGCGGAGAUUUCUCGUUUCUCGCCGAGAAUCCCGCCAGCGC ((((((........(((((((.(((((((((............((((((.........)))))).((((...-.)))).....))))))))).))))))))))))).............. ( -42.00) >DroSim_CAF1 15866 119 - 1 CUCGGCCAUAAACCGAGAAACAAGAAAUUUCAAUUUCAAAUUUAAAGCGUCUAACUUUCGCUUUCCUCCGCA-AGGAGAUGCGGAGAUUUCUCGUUUCUCGCCGAGAAUCCCGCCAGCGC ((((((........(((((((.(((((((((............((((((.........)))))).((((...-.)))).....))))))))).))))))))))))).............. ( -42.00) >DroEre_CAF1 10154 119 - 1 CUCGGCCAUAAGCCGAGAAACAAGAAAUUUCAAUUUUAAAUUUAAAGCGUCUAACUUUCGCUUUCCUCCGCA-AGGAGAUGCGGAGAUUGCACGUUUCUCGCCGAGAAUCCCGCCUGCAU ((((((........(((((((....((((((............((((((.........)))))).((((...-.)))).....))))))....))))))))))))).............. ( -35.80) >DroYak_CAF1 21605 120 - 1 CUCGGCCAUAAACCGAGUAACAAGAAAUUUCAAUUUCAAAUUUAAAGCGUCUAACUUUCGCUUUCCUCCGCAAAGGAGAUGCGGAGAUUUCUCGUUUCUCGCCAAGAAUCCCGCCUGCAC ...(((.......((((.(((.(((((((((............((((((.........)))))).((((.....)))).....))))))))).))).))))....(.....))))..... ( -30.20) >consensus CUCGGCCAUAAACCGAGAAACAAGAAAUUUCAAUUUCAAAUUUAAAGCGUCUAACUUUCGCUUUCCUCCGCA_AGGAGAUGCGGAGAUUUCUCGUUUCUCGCCGAGAAUCCCGCCUGCGC ((((((........(((((((.(((((((((............((((((.........)))))).((((.....)))).....))))))))).))))))))))))).............. (-32.84 = -34.04 + 1.20)

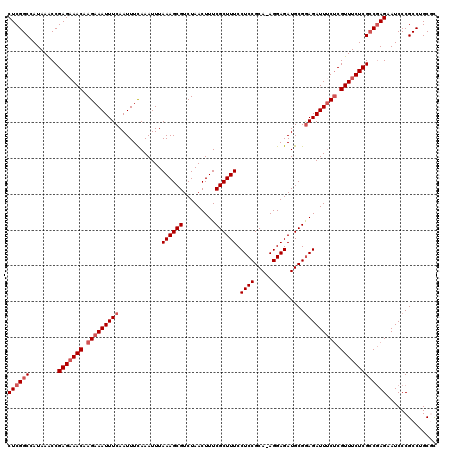
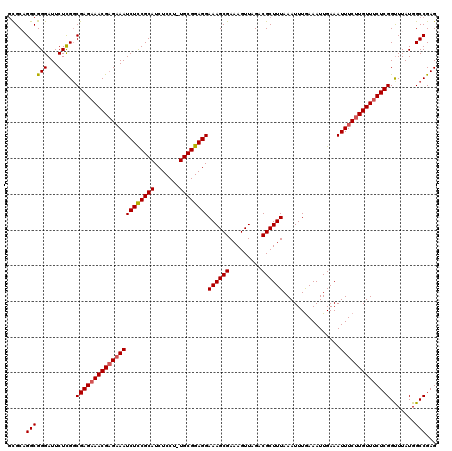
| Location | 17,656,410 – 17,656,527 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.04 |
| Mean single sequence MFE | -33.10 |
| Consensus MFE | -29.76 |
| Energy contribution | -30.00 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627154 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17656410 117 + 20766785 AUCUCCU-UGCGGAGGAAAGCGAAAGUUAGACGCUUUAAAUUUGAAAUUGAAAUUUCUUGUUUCUCGGUUUAUGGCCGAGGAAAAGCAAAAGCCGCGGUUC--GAGACUCCAGCCAAGUG .((((.(-(((((.....(((....)))....(((((......(((((....)))))....((((((((.....))))))))))))).....))))))...--))))............. ( -32.10) >DroSec_CAF1 9953 117 + 1 AUCUCCU-UGCGGAGGAAAGCGAAAGUUAGACGCUUUAAAUUUGAAAUUGAAAUUUCUUGUUUCUCGGUUUAUGGCCGAGGAAAAGCAAAAGCCGCGGUUC--GAGACUCCAGCCAAGUG .((((.(-(((((.....(((....)))....(((((......(((((....)))))....((((((((.....))))))))))))).....))))))...--))))............. ( -32.10) >DroSim_CAF1 15906 117 + 1 AUCUCCU-UGCGGAGGAAAGCGAAAGUUAGACGCUUUAAAUUUGAAAUUGAAAUUUCUUGUUUCUCGGUUUAUGGCCGAGGAAAAGCAAAAGCCGCGGCUC--GAGACUCCAGCCAAGUG .......-.((((.....(((....)))....(((((......(((((....)))))....((((((((.....))))))))))))).....))))((((.--(.....).))))..... ( -34.30) >DroEre_CAF1 10194 117 + 1 AUCUCCU-UGCGGAGGAAAGCGAAAGUUAGACGCUUUAAAUUUAAAAUUGAAAUUUCUUGUUUCUCGGCUUAUGGCCGAGGAAAAGCAAAAGCCGCGUUUC--GAGACUCCAGCCAAGUG .....((-(((((((...(((....)))((((((..........................(((((((((.....)))))))))..((....)).)))))).--....)))).).)))).. ( -33.30) >DroYak_CAF1 21645 120 + 1 AUCUCCUUUGCGGAGGAAAGCGAAAGUUAGACGCUUUAAAUUUGAAAUUGAAAUUUCUUGUUACUCGGUUUAUGGCCGAGGAAAAGCAAAAGCCGCGAUUCGCGAGACUCCAGCCAAGUG .....(((.((((((...(((....)))....(((((.(((..(((((....)))))..))).((((((.....))))))..)))))......((((...))))...)))).)).))).. ( -33.70) >consensus AUCUCCU_UGCGGAGGAAAGCGAAAGUUAGACGCUUUAAAUUUGAAAUUGAAAUUUCUUGUUUCUCGGUUUAUGGCCGAGGAAAAGCAAAAGCCGCGGUUC__GAGACUCCAGCCAAGUG .((((...(((((.....(((....)))....(((((......(((((....)))))....((((((((.....))))))))))))).....)))))......))))............. (-29.76 = -30.00 + 0.24)



| Location | 17,656,410 – 17,656,527 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.04 |
| Mean single sequence MFE | -32.86 |
| Consensus MFE | -27.86 |
| Energy contribution | -27.86 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.989219 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17656410 117 - 20766785 CACUUGGCUGGAGUCUC--GAACCGCGGCUUUUGCUUUUCCUCGGCCAUAAACCGAGAAACAAGAAAUUUCAAUUUCAAAUUUAAAGCGUCUAACUUUCGCUUUCCUCCGCA-AGGAGAU ..((((((..(((((.(--(...)).)))))..)).....(((((.......)))))...))))...................((((((.........)))))).((((...-.)))).. ( -32.60) >DroSec_CAF1 9953 117 - 1 CACUUGGCUGGAGUCUC--GAACCGCGGCUUUUGCUUUUCCUCGGCCAUAAACCGAGAAACAAGAAAUUUCAAUUUCAAAUUUAAAGCGUCUAACUUUCGCUUUCCUCCGCA-AGGAGAU ..((((((..(((((.(--(...)).)))))..)).....(((((.......)))))...))))...................((((((.........)))))).((((...-.)))).. ( -32.60) >DroSim_CAF1 15906 117 - 1 CACUUGGCUGGAGUCUC--GAGCCGCGGCUUUUGCUUUUCCUCGGCCAUAAACCGAGAAACAAGAAAUUUCAAUUUCAAAUUUAAAGCGUCUAACUUUCGCUUUCCUCCGCA-AGGAGAU .....((((.(.....)--.))))((((....((((((..(((((.......)))))......(((((....)))))......))))))........))))....((((...-.)))).. ( -32.80) >DroEre_CAF1 10194 117 - 1 CACUUGGCUGGAGUCUC--GAAACGCGGCUUUUGCUUUUCCUCGGCCAUAAGCCGAGAAACAAGAAAUUUCAAUUUUAAAUUUAAAGCGUCUAACUUUCGCUUUCCUCCGCA-AGGAGAU ..((((((..(((((.(--(...)).)))))..)).....((((((.....))))))...))))...................((((((.........)))))).((((...-.)))).. ( -36.20) >DroYak_CAF1 21645 120 - 1 CACUUGGCUGGAGUCUCGCGAAUCGCGGCUUUUGCUUUUCCUCGGCCAUAAACCGAGUAACAAGAAAUUUCAAUUUCAAAUUUAAAGCGUCUAACUUUCGCUUUCCUCCGCAAAGGAGAU ..((((((..((((..((((...))))))))..)).....(((((.......)))))...))))...................((((((.........)))))).((((.....)))).. ( -30.10) >consensus CACUUGGCUGGAGUCUC__GAACCGCGGCUUUUGCUUUUCCUCGGCCAUAAACCGAGAAACAAGAAAUUUCAAUUUCAAAUUUAAAGCGUCUAACUUUCGCUUUCCUCCGCA_AGGAGAU ..((((((..(((((...........)))))..)).....(((((.......)))))...))))...................((((((.........)))))).((((.....)))).. (-27.86 = -27.86 + -0.00)

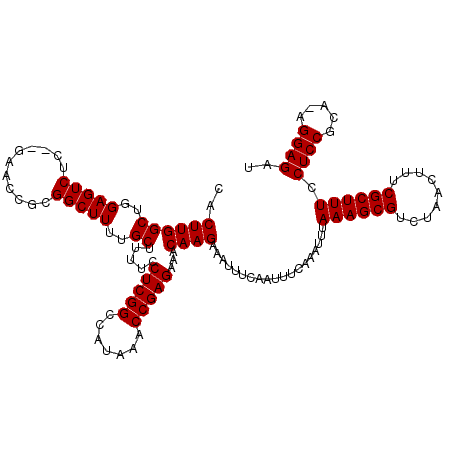
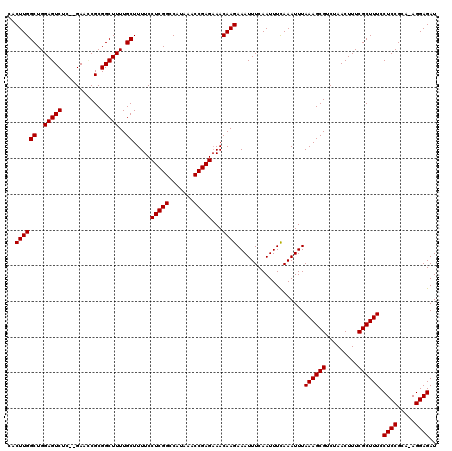
| Location | 17,656,449 – 17,656,563 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.45 |
| Mean single sequence MFE | -35.52 |
| Consensus MFE | -30.82 |
| Energy contribution | -31.14 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.644979 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17656449 114 + 20766785 UUUGAAAUUGAAAUUUCUUGUUUCUCGGUUUAUGGCCGAGGAAAAGCAAAAGCCGCGGUUC--GAGACUCCAGCCAAGUGCACGUU-CGGUUUGGGCCAU---AAACUGUGGCCUGCCAU ....((((((((.((((((.(((((((((.....)))))))))))).))).(((..((((.--(.....).))))..).))...))-))))))(((((((---.....)))))))..... ( -34.30) >DroSec_CAF1 9992 117 + 1 UUUGAAAUUGAAAUUUCUUGUUUCUCGGUUUAUGGCCGAGGAAAAGCAAAAGCCGCGGUUC--GAGACUCCAGCCAAGUGCACGUU-CGGUUUGGGCCAUAAUAAACUGUGGCCUGCCAU ....((((((((.((((((.(((((((((.....)))))))))))).))).(((..((((.--(.....).))))..).))...))-))))))((((((((......))))))))..... ( -35.50) >DroSim_CAF1 15945 114 + 1 UUUGAAAUUGAAAUUUCUUGUUUCUCGGUUUAUGGCCGAGGAAAAGCAAAAGCCGCGGCUC--GAGACUCCAGCCAAGUGCACGUU-CGGUUUGGGCCAU---AAACUGUGGCCUGCCAU ....((((((((.((((((.(((((((((.....)))))))))))).))).(((..((((.--(.....).))))..).))...))-))))))(((((((---.....)))))))..... ( -36.70) >DroEre_CAF1 10233 114 + 1 UUUAAAAUUGAAAUUUCUUGUUUCUCGGCUUAUGGCCGAGGAAAAGCAAAAGCCGCGUUUC--GAGACUCCAGCCAAGUGCACGUU-UGGUUUGGGCCAU---AAACUGUGGCCUGCCAU .......(((((((......(((((((((.....)))))))))..((.......)))))))--))......(((((((......))-))))).(((((((---.....)))))))..... ( -34.90) >DroYak_CAF1 21685 117 + 1 UUUGAAAUUGAAAUUUCUUGUUACUCGGUUUAUGGCCGAGGAAAAGCAAAAGCCGCGAUUCGCGAGACUCCAGCCAAGUGCACGUUUUGGUUUGGGCCAU---AAACUGUGGCCUGCCAU ...(((((....)))))........((((((((((((..(((...((....))((((...))))....)))(((((((.......)))))))..))))))---))))))(((....))). ( -36.20) >consensus UUUGAAAUUGAAAUUUCUUGUUUCUCGGUUUAUGGCCGAGGAAAAGCAAAAGCCGCGGUUC__GAGACUCCAGCCAAGUGCACGUU_CGGUUUGGGCCAU___AAACUGUGGCCUGCCAU ...(((((....)))))...(((((((((.....)))))))))..(((...((.(.((.((....)).))).))....))).......(((..(((((((........)))))))))).. (-30.82 = -31.14 + 0.32)

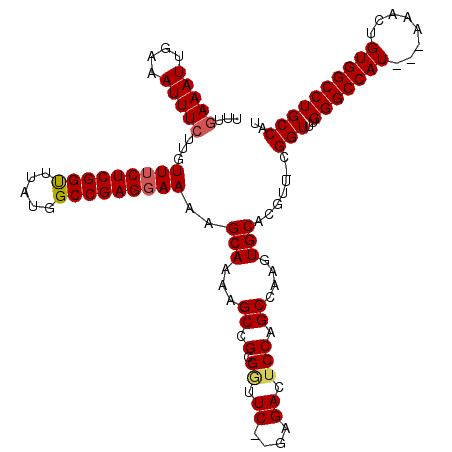
| Location | 17,656,489 – 17,656,603 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.80 |
| Mean single sequence MFE | -42.44 |
| Consensus MFE | -35.29 |
| Energy contribution | -36.09 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.932991 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17656489 114 + 20766785 GAAAAGCAAAAGCCGCGGUUC--GAGACUCCAGCCAAGUGCACGUU-CGGUUUGGGCCAU---AAACUGUGGCCUGCCAUUAUGAGCUAACUUGGCCAGGGCGGCGAAGAAGGGGUUUCU (((..((.......))..)))--((((((((...(((((((.(((.-.(((..(((((((---.....))))))))))...))).))..)))))(((.....)))......)))))))). ( -42.30) >DroSec_CAF1 10032 114 + 1 GAAAAGCAAAAGCCGCGGUUC--GAGACUCCAGCCAAGUGCACGUU-CGGUUUGGGCCAUAAUAAACUGUGGCCUGCCAUUAUGAGCUAACUUGGCCAGGGCGGCGGAGA---GGUUUCU ((((.......(((((((((.--..))))((.(((((((((.(((.-.(((..((((((((......)))))))))))...))).))..)))))))..))))))).....---..)))). ( -43.44) >DroSim_CAF1 15985 111 + 1 GAAAAGCAAAAGCCGCGGCUC--GAGACUCCAGCCAAGUGCACGUU-CGGUUUGGGCCAU---AAACUGUGGCCUGCCAUUAUGAGCUAACUUGGCCAGGGCGGCGGAGA---GGUUUCU ...((((.....((((.((((--(......).(((((((((.(((.-.(((..(((((((---.....))))))))))...))).))..)))))))..)))).))))...---.)))).. ( -43.80) >DroEre_CAF1 10273 111 + 1 GAAAAGCAAAAGCCGCGUUUC--GAGACUCCAGCCAAGUGCACGUU-UGGUUUGGGCCAU---AAACUGUGGCCUGCCAUUAUGAGCUAACGUGGCCAGGGCGGCGGAGA---GGUUUCU ((((.......((((((((..--..))).((.((((.((((.(((.-((((..(((((((---.....)))))))))))..))).))..)).))))..))))))).....---..)))). ( -40.54) >DroYak_CAF1 21725 114 + 1 GAAAAGCAAAAGCCGCGAUUCGCGAGACUCCAGCCAAGUGCACGUUUUGGUUUGGGCCAU---AAACUGUGGCCUGCCAUUAUGAGCUAACGCGGCCAGGGCGACGGAGA---GGUUUCU .........((((((((...)))....((((.(((..(.((.(((..((((..(((((((---.....)))))))))))..))).((....)).)))..)))...)))).---))))).. ( -42.10) >consensus GAAAAGCAAAAGCCGCGGUUC__GAGACUCCAGCCAAGUGCACGUU_CGGUUUGGGCCAU___AAACUGUGGCCUGCCAUUAUGAGCUAACUUGGCCAGGGCGGCGGAGA___GGUUUCU ((((.......(((((...((....))..((.(((((((((.(((...(((..(((((((........))))))))))...))).))..)))))))..)))))))..........)))). (-35.29 = -36.09 + 0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:47:09 2006