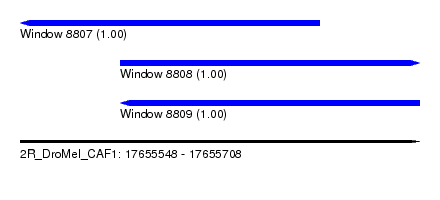
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,655,548 – 17,655,708 |
| Length | 160 |
| Max. P | 0.999841 |

| Location | 17,655,548 – 17,655,668 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.17 |
| Mean single sequence MFE | -44.81 |
| Consensus MFE | -39.88 |
| Energy contribution | -39.60 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.75 |
| SVM RNA-class probability | 0.999586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17655548 120 - 20766785 AACAAUUUCCACGUCAGGGAAGCGUAGGCAGAGUUCGACUAUAUCGCAGGUCUAUUGACGUUUGUCUGAGCUCUGUCUACGCAGCUGGAUGUCUGUUAACCCCUCCGAGGCCCCACGAAU ...........(((..(((..(((((((((((((((((((........))))....(((....))).))))))))))))))).((((((.(..(....)..).)))..)))))))))... ( -42.30) >DroSec_CAF1 9112 120 - 1 GACAAUUUCCACGUCAGGGUAGCGUAGGCAGAGUUCGACUAUAUCGCAGCUCUAUUGACGUUUGUCUGAGCUCUGUCUACGCAGCUGGAUGCCUGUUAACCCCUCCGAGGCCCCACAAAU (((.........))).(((..((((((((((((((((((.....(((((.....))).))...))).)))))))))))))))........((((.............)))))))...... ( -41.72) >DroSim_CAF1 15048 120 - 1 GACAAUUUCCACGUCAGGGUAGCGUAGGCAGAGUUCGACUAUAUCGCAGCUCUAUUGACGUUUGUCUGAGCUCUGUCUACGCAGCUGGAUGCCUGUUAACCCCUCCGAGGCCCCACGAAU (((.........))).(((..((((((((((((((((((.....(((((.....))).))...))).)))))))))))))))........((((.............)))))))...... ( -41.72) >DroEre_CAF1 9330 120 - 1 GGCCAUUUUCUGGCCAGGGAAGCGUAGGCAGAGUUCGACUAUAUCGCAGGUCUAUUGACGUUUGUCUGAGCUCUGUCUACGCAGCUGGAUGUCUGGUAACCCCUCCGAAGCCCCACGGAU (((((.....))))).(((..(((((((((((((((((((........))))....(((....))).))))))))))))))).((..(....)..))...)))((((........)))). ( -53.80) >DroYak_CAF1 20777 120 - 1 GAUAAUUUCCACGUCAGGGAAGCGUAGGCAGAGUUCGACUAUAUUGCUGGUCUAUUGACGUUUGUCUGAGCUCUGUCUACGCAGCUGGAUGUCUGGUAACCCCGCCGAAGCCCCACGAAU ...........((.(.(((..((((((((((((((((((((......)))))....(((....))).))))))))))))))).((..(....)..))..))).).))............. ( -44.50) >consensus GACAAUUUCCACGUCAGGGAAGCGUAGGCAGAGUUCGACUAUAUCGCAGGUCUAUUGACGUUUGUCUGAGCUCUGUCUACGCAGCUGGAUGUCUGUUAACCCCUCCGAGGCCCCACGAAU (((.........))).(((..((((((((((((((((((.....(((((.....))).))...))).))))))))))))))).((((((.(..(....)..).)))..))))))...... (-39.88 = -39.60 + -0.28)



| Location | 17,655,588 – 17,655,708 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.17 |
| Mean single sequence MFE | -35.26 |
| Consensus MFE | -33.50 |
| Energy contribution | -33.38 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.74 |
| SVM RNA-class probability | 0.996745 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17655588 120 + 20766785 GUAGACAGAGCUCAGACAAACGUCAAUAGACCUGCGAUAUAGUCGAACUCUGCCUACGCUUCCCUGACGUGGAAAUUGUUGGGAAAUUACCGGGAAGUGGCAAACGAAAGCAGUUUGAAC ((((.(((((.((.(((....)))....(((..........))))).))))).))))(((((((.((((.......))))((.......)))))))))..(((((.......)))))... ( -35.30) >DroSec_CAF1 9152 120 + 1 GUAGACAGAGCUCAGACAAACGUCAAUAGAGCUGCGAUAUAGUCGAACUCUGCCUACGCUACCCUGACGUGGAAAUUGUCGGGAAAUUACCGGGAAGUGGCAAACGAAAGCAGUUUGAAC ((((.((((((((.(((....)))....))))..((((...))))...)))).))))(((.(((.((((.......))))((.......))))).)))..(((((.......)))))... ( -33.00) >DroSim_CAF1 15088 120 + 1 GUAGACAGAGCUCAGACAAACGUCAAUAGAGCUGCGAUAUAGUCGAACUCUGCCUACGCUACCCUGACGUGGAAAUUGUCGGGAAAUUACCGGGAAGUGGCAAACGAAAGCAGUUUGAAC ((((.((((((((.(((....)))....))))..((((...))))...)))).))))(((.(((.((((.......))))((.......))))).)))..(((((.......)))))... ( -33.00) >DroEre_CAF1 9370 120 + 1 GUAGACAGAGCUCAGACAAACGUCAAUAGACCUGCGAUAUAGUCGAACUCUGCCUACGCUUCCCUGGCCAGAAAAUGGCCGGGAAAUUACCGGGAAGUGGCAAGCGAAAGCAGUUUGAAU ((((.(((((.((.(((....)))....(((..........))))).))))).))))(((((((.(((((.....)))))((.......))))))))).....((....))......... ( -45.60) >DroYak_CAF1 20817 119 + 1 GUAGACAGAGCUCAGACAAACGUCAAUAGACCAGCAAUAUAGUCGAACUCUGCCUACGCUUCCCUGACGUGGAAAUUAUCGAGAAAUUACCGGGAAGUG-CAAAUAAAAGCGGUUUGAAU ((((.(((((.((.(((....)))....(((.(......).))))).))))).))))(((((((......((.((((.......)))).))))))))).-(((((.......)))))... ( -29.40) >consensus GUAGACAGAGCUCAGACAAACGUCAAUAGACCUGCGAUAUAGUCGAACUCUGCCUACGCUUCCCUGACGUGGAAAUUGUCGGGAAAUUACCGGGAAGUGGCAAACGAAAGCAGUUUGAAC ((((.(((((.((.(((....)))....((.(((.....))))))).))))).))))(((((((.((((.......))))((.......)))))))))..(((((.......)))))... (-33.50 = -33.38 + -0.12)

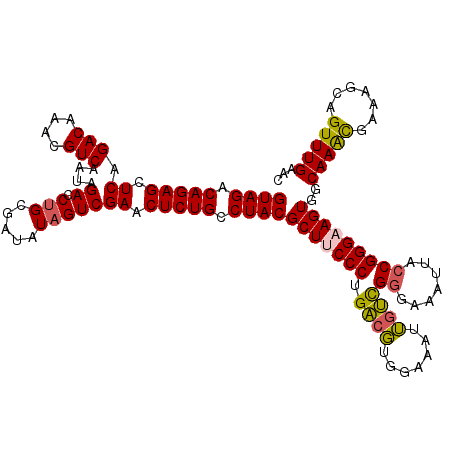
| Location | 17,655,588 – 17,655,708 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.17 |
| Mean single sequence MFE | -38.48 |
| Consensus MFE | -34.50 |
| Energy contribution | -34.70 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.90 |
| SVM decision value | 4.22 |
| SVM RNA-class probability | 0.999841 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17655588 120 - 20766785 GUUCAAACUGCUUUCGUUUGCCACUUCCCGGUAAUUUCCCAACAAUUUCCACGUCAGGGAAGCGUAGGCAGAGUUCGACUAUAUCGCAGGUCUAUUGACGUUUGUCUGAGCUCUGUCUAC .........((......(((((.......))))).(((((................)))))))(((((((((((((((((........))))....(((....))).))))))))))))) ( -35.99) >DroSec_CAF1 9152 120 - 1 GUUCAAACUGCUUUCGUUUGCCACUUCCCGGUAAUUUCCCGACAAUUUCCACGUCAGGGUAGCGUAGGCAGAGUUCGACUAUAUCGCAGCUCUAUUGACGUUUGUCUGAGCUCUGUCUAC .........(((.....(((((.......)))))...((((((.........))).))).)))((((((((((((((((.....(((((.....))).))...))).))))))))))))) ( -36.20) >DroSim_CAF1 15088 120 - 1 GUUCAAACUGCUUUCGUUUGCCACUUCCCGGUAAUUUCCCGACAAUUUCCACGUCAGGGUAGCGUAGGCAGAGUUCGACUAUAUCGCAGCUCUAUUGACGUUUGUCUGAGCUCUGUCUAC .........(((.....(((((.......)))))...((((((.........))).))).)))((((((((((((((((.....(((((.....))).))...))).))))))))))))) ( -36.20) >DroEre_CAF1 9370 120 - 1 AUUCAAACUGCUUUCGCUUGCCACUUCCCGGUAAUUUCCCGGCCAUUUUCUGGCCAGGGAAGCGUAGGCAGAGUUCGACUAUAUCGCAGGUCUAUUGACGUUUGUCUGAGCUCUGUCUAC ...............(((((((.......))))).((((((((((.....))))).)))))))(((((((((((((((((........))))....(((....))).))))))))))))) ( -47.10) >DroYak_CAF1 20817 119 - 1 AUUCAAACCGCUUUUAUUUG-CACUUCCCGGUAAUUUCUCGAUAAUUUCCACGUCAGGGAAGCGUAGGCAGAGUUCGACUAUAUUGCUGGUCUAUUGACGUUUGUCUGAGCUCUGUCUAC .........((........)-).((((((((.((((.......)))).))......)))))).((((((((((((((((((......)))))....(((....))).))))))))))))) ( -36.90) >consensus GUUCAAACUGCUUUCGUUUGCCACUUCCCGGUAAUUUCCCGACAAUUUCCACGUCAGGGAAGCGUAGGCAGAGUUCGACUAUAUCGCAGGUCUAUUGACGUUUGUCUGAGCUCUGUCUAC .........((......(((((.......))))).((((((((.........))).)))))))((((((((((((((((.....(((((.....))).))...))).))))))))))))) (-34.50 = -34.70 + 0.20)

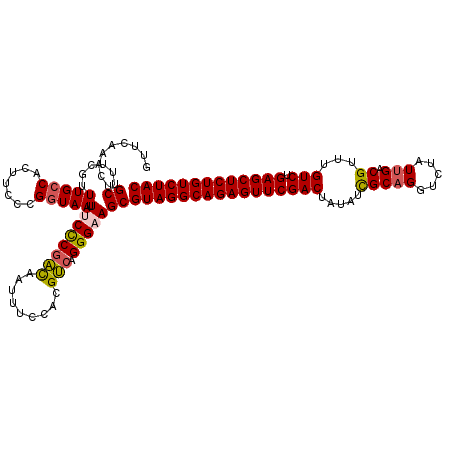
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:59 2006