
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,655,076 – 17,655,413 |
| Length | 337 |
| Max. P | 0.991872 |

| Location | 17,655,076 – 17,655,189 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.54 |
| Mean single sequence MFE | -28.42 |
| Consensus MFE | -26.00 |
| Energy contribution | -25.72 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.983098 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17655076 113 - 20766785 CAGGUGGCCGCUUUGGAUUUCUAAUAGUUUCGACUGCCGUAGCGGUGGUUCUCUGUUUGAAAACGGAAGCAAAGCCCAAACAAAAAAUGAAAAUAAACACCAA-------ACUCACAUCC ..((((...(((((.(.((((.(((((....(((..(((...)))..)))..))))).)))).).)))))................((....))...))))..-------.......... ( -28.60) >DroSec_CAF1 8635 113 - 1 CAGGUGGCCGCUUUGGAUUUCUAAUAGUUUCGACCGCCGUAGCGGUGGUUCUCUGUUCGAAAACGGAAGCAAAGCCCAAACAAAAAAUGAAAAUAAACACCAA-------ACUCACAGCC ..((((...(((((.(.((((.(((((....((((((((...))))))))..))))).)))).).)))))................((....))...))))..-------.......... ( -31.40) >DroSim_CAF1 14566 113 - 1 CAGGUGGCCGCUUUGGAUUUCUAAUAGUUUCGAACGCCGUAGCGGUGGUUCUCUGUUCGAAAACGGAAGCAAAGCCCAAACAAAAAAUGAAAAUAAACACCAA-------ACUCACAGCC ..((((((((((.(((..(((.((....)).)))..))).)))))).(((.((((((....)))))))))...........................))))..-------.......... ( -26.70) >DroEre_CAF1 8871 113 - 1 CAGGUGGCUGCUUUUUAUUUCUAAUGGUUUAGACCGCCGUAGCGGUGGUUCCCUGUUUGAAAACGGAAGCAAAGCCCAAACAAGAAAUAAAAAUAAACACCAA-------ACUCACAUCC ..((((..((.(((((((((((...(((((.((((((((...))))))))..(((((....))))).....)))))......)))))))))))))..))))..-------.......... ( -32.30) >DroYak_CAF1 20305 120 - 1 CAGGUGGCAGCUUUGGAUUUCUAAUGGUUUCGAUCGUCGUAGCGGUGGUUCUCUGUUUGAAAACGGAAGCAAAGCCCAAACAAAAAAUACAAAUAAACACCAAACACAAAACUCACAUCC ..((((...(((((.(.((((.(((((...(.(((((....))))).)....))))).)))).).)))))...(......)................))))................... ( -23.10) >consensus CAGGUGGCCGCUUUGGAUUUCUAAUAGUUUCGACCGCCGUAGCGGUGGUUCUCUGUUUGAAAACGGAAGCAAAGCCCAAACAAAAAAUGAAAAUAAACACCAA_______ACUCACAUCC ..((((...(((((.(.((((.(((((....((((((((...))))))))..))))).)))).).)))))................((....))...))))................... (-26.00 = -25.72 + -0.28)

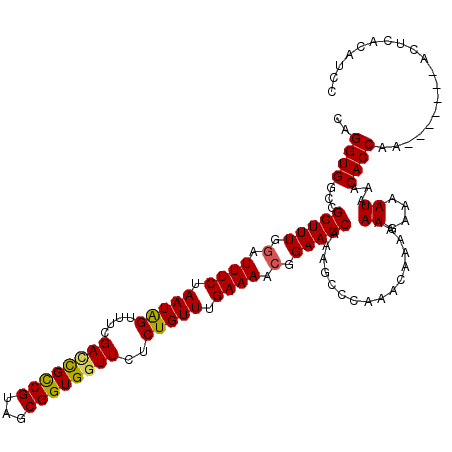
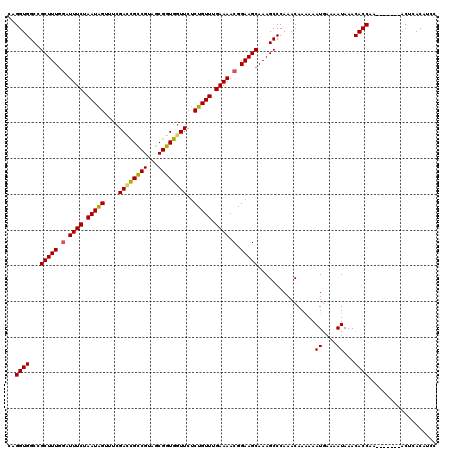
| Location | 17,655,109 – 17,655,223 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.44 |
| Mean single sequence MFE | -35.58 |
| Consensus MFE | -29.78 |
| Energy contribution | -29.90 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.686629 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17655109 114 - 20766785 UGAUU------GUGCACUUGCCACAGAAAAUCGCCGUCUACAGGUGGCCGCUUUGGAUUUCUAAUAGUUUCGACUGCCGUAGCGGUGGUUCUCUGUUUGAAAACGGAAGCAAAGCCCAAA ...((------(.((..((((..((((.((((((((.((((.((..(.((((((((....)))).))...)).)..)))))))))))))).))))((((....)))).)))).)).))). ( -34.90) >DroSec_CAF1 8668 114 - 1 UGAUU------GUGCACUUGCCACAGAAAAUCGCCGACUACAGGUGGCCGCUUUGGAUUUCUAAUAGUUUCGACCGCCGUAGCGGUGGUUCUCUGUUCGAAAACGGAAGCAAAGCCCAAA ...((------(.((..((((..((((.((((((((.((((.(((((.((((((((....)))).))...)).))))))))))))))))).))))((((....)))).)))).)).))). ( -39.70) >DroSim_CAF1 14599 114 - 1 UGAUU------GUGCACUUGCCACAGAAAAUCGCCGACUACAGGUGGCCGCUUUGGAUUUCUAAUAGUUUCGAACGCCGUAGCGGUGGUUCUCUGUUCGAAAACGGAAGCAAAGCCCAAA ...((------(.((..((((..((((.((((((((.((((.((((..((((((((....)))).))...))..)))))))))))))))).))))((((....)))).)))).)).))). ( -37.60) >DroEre_CAF1 8904 114 - 1 UGAUU------GUGCACUUGCCAUAGAAAAACGCCGACUACAGGUGGCUGCUUUUUAUUUCUAAUGGUUUAGACCGCCGUAGCGGUGGUUCCCUGUUUGAAAACGGAAGCAAAGCCCAAA ...((------(.((..((((....(((...(((((.((((.((((((((((..(((....))).)))..)).)))))))))))))).))).(((((....)))))..)))).)).))). ( -32.70) >DroYak_CAF1 20345 120 - 1 UGAUUGCAUUUGUGCACUUGCCACAGAAAAACGCCGACUACAGGUGGCAGCUUUGGAUUUCUAAUGGUUUCGAUCGUCGUAGCGGUGGUUCUCUGUUUGAAAACGGAAGCAAAGCCCAAA ...((((.(((((((....).))))))....(((((.((((.(((.(.((((((((....)))).)))).).)))...)))))))))....((((((....)))))).))))........ ( -33.00) >consensus UGAUU______GUGCACUUGCCACAGAAAAUCGCCGACUACAGGUGGCCGCUUUGGAUUUCUAAUAGUUUCGACCGCCGUAGCGGUGGUUCUCUGUUUGAAAACGGAAGCAAAGCCCAAA ...........(.((..((((..((((.((((((((.((((.((((((.(((((((....)))).)))...).))))))))))))))))).))))((((....)))).)))).)).)... (-29.78 = -29.90 + 0.12)

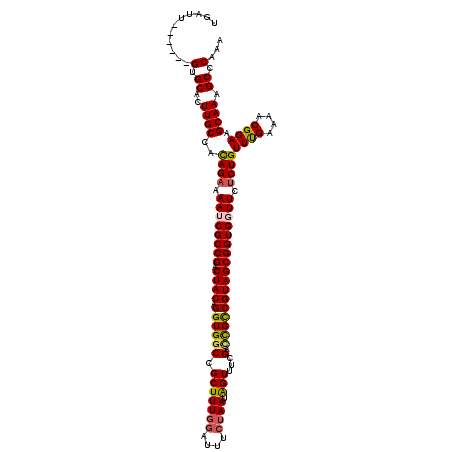
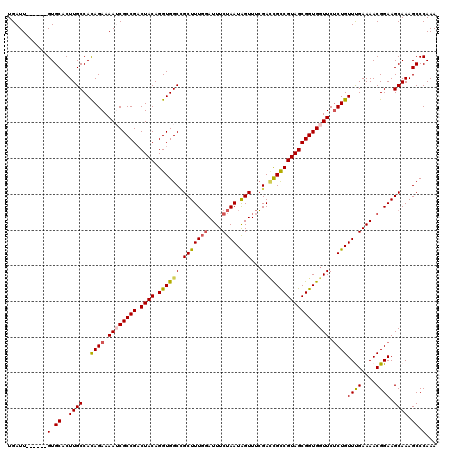
| Location | 17,655,189 – 17,655,303 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.59 |
| Mean single sequence MFE | -31.44 |
| Consensus MFE | -21.36 |
| Energy contribution | -22.32 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.544204 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17655189 114 + 20766785 UAGACGGCGAUUUUCUGUGGCAAGUGCAC------AAUCAACUGCAAUUGCAGUGGCAAUAGCAAUAGCAACCGCGUCGUUUCCGCUGCUCUUGGCCAAAAAAGGCAUUCGACAAUUAUU .((((((((......(((((....(((..------.....(((((....))))).((....))....))).))))).......))))).)))..(((......))).............. ( -28.52) >DroSec_CAF1 8748 114 + 1 UAGUCGGCGAUUUUCUGUGGCAAGUGCAC------AAUCAACUGCAAUUGCAGUGGCAACAGCAAUAGCAACCGCGUCGUUUCCGCUGCUCUUGGCCAAAAAAGGCAUUCGACAAUUAUU ..(((((...((((...(((((((.(((.------.......(((.(((((..((....))))))).)))...(((.......)))))).))).))))..))))....)))))....... ( -31.40) >DroSim_CAF1 14679 114 + 1 UAGUCGGCGAUUUUCUGUGGCAAGUGCAC------AAUCAACUGCAAUUGCAGUGGCAACAGCAAUAGCAACCGCGUCGUUUCCGCUGCUCUUGGCCAAAAAAGGCAUUCGACAAUUAUU ..(((((...((((...(((((((.(((.------.......(((.(((((..((....))))))).)))...(((.......)))))).))).))))..))))....)))))....... ( -31.40) >DroEre_CAF1 8984 96 + 1 UAGUCGGCGUUUUUCUAUGGCAAGUGCAC------AAUCAACUGCAAUUGC------A------------ACCGCAGCAUUUCCGCUGCUUUUGGCCAAAAAAGGCAUUCGACAAUUAUU ..((((((.(((((...((((...((((.------.......))))....(------(------------(..(((((......)))))..)))))))))))).))...))))....... ( -27.90) >DroYak_CAF1 20425 108 + 1 UAGUCGGCGUUUUUCUGUGGCAAGUGCACAAAUGCAAUCAACUGCAAUUGCAGUGGCA------------AUUGCAGCGUUUCCGCUGCUUUUGGCCAAAAAAGGCAUUCGACAAUUAUU ..(((((.((((((...((((...((((....))))....(((((....)))))..((------------(..((((((....))))))..)))))))..))))))..)))))....... ( -38.00) >consensus UAGUCGGCGAUUUUCUGUGGCAAGUGCAC______AAUCAACUGCAAUUGCAGUGGCAA_AGCAAUAGCAACCGCGUCGUUUCCGCUGCUCUUGGCCAAAAAAGGCAUUCGACAAUUAUU ..(((((.............((((.(((............(((((....)))))...................(((.......)))))).))))(((......)))..)))))....... (-21.36 = -22.32 + 0.96)

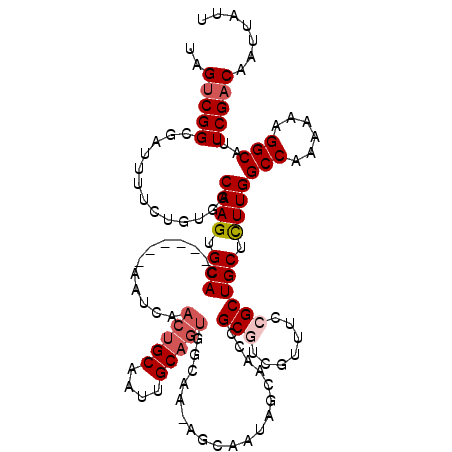
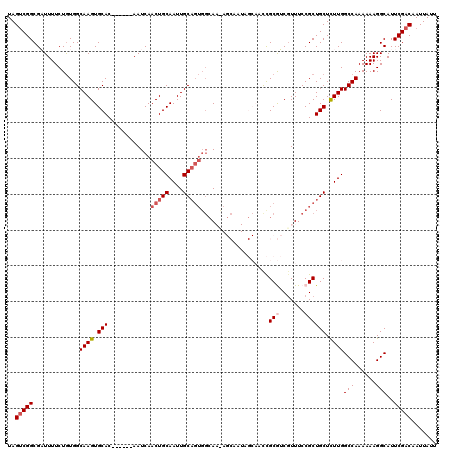
| Location | 17,655,189 – 17,655,303 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.59 |
| Mean single sequence MFE | -32.82 |
| Consensus MFE | -23.06 |
| Energy contribution | -24.72 |
| Covariance contribution | 1.66 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.644902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17655189 114 - 20766785 AAUAAUUGUCGAAUGCCUUUUUUGGCCAAGAGCAGCGGAAACGACGCGGUUGCUAUUGCUAUUGCCACUGCAAUUGCAGUUGAUU------GUGCACUUGCCACAGAAAAUCGCCGUCUA ..........((..((.((((((((.((((.((((((....)).)((((((((....((....))....))))))))........------.))).)))).).)))))))..))..)).. ( -31.70) >DroSec_CAF1 8748 114 - 1 AAUAAUUGUCGAAUGCCUUUUUUGGCCAAGAGCAGCGGAAACGACGCGGUUGCUAUUGCUGUUGCCACUGCAAUUGCAGUUGAUU------GUGCACUUGCCACAGAAAAUCGCCGACUA .......((((...((.((((((((.((((.((((((....)).)((((((((....((....))....))))))))........------.))).)))).).)))))))..)))))).. ( -34.60) >DroSim_CAF1 14679 114 - 1 AAUAAUUGUCGAAUGCCUUUUUUGGCCAAGAGCAGCGGAAACGACGCGGUUGCUAUUGCUGUUGCCACUGCAAUUGCAGUUGAUU------GUGCACUUGCCACAGAAAAUCGCCGACUA .......((((...((.((((((((.((((.((((((....)).)((((((((....((....))....))))))))........------.))).)))).).)))))))..)))))).. ( -34.60) >DroEre_CAF1 8984 96 - 1 AAUAAUUGUCGAAUGCCUUUUUUGGCCAAAAGCAGCGGAAAUGCUGCGGU------------U------GCAAUUGCAGUUGAUU------GUGCACUUGCCAUAGAAAAACGCCGACUA .......((((...((.((((((((((....((((((....)))))))))------------)------((((.((((.......------.)))).))))....)))))).)))))).. ( -26.60) >DroYak_CAF1 20425 108 - 1 AAUAAUUGUCGAAUGCCUUUUUUGGCCAAAAGCAGCGGAAACGCUGCAAU------------UGCCACUGCAAUUGCAGUUGAUUGCAUUUGUGCACUUGCCACAGAAAAACGCCGACUA .......((((...((.(((((((((.....((((((....))))))...------------(((((.(((((((......)))))))..)).)))...))))...))))).)))))).. ( -36.60) >consensus AAUAAUUGUCGAAUGCCUUUUUUGGCCAAGAGCAGCGGAAACGACGCGGUUGCUAUUGCU_UUGCCACUGCAAUUGCAGUUGAUU______GUGCACUUGCCACAGAAAAUCGCCGACUA .......((((...((.((((((((.((((.(((.((....))..((((((((....((....))....))))))))...............))).)))).).)))))))..)))))).. (-23.06 = -24.72 + 1.66)


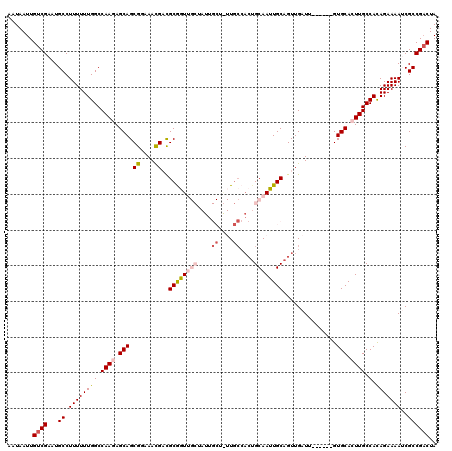
| Location | 17,655,223 – 17,655,343 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.31 |
| Mean single sequence MFE | -34.92 |
| Consensus MFE | -21.24 |
| Energy contribution | -23.05 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.622719 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17655223 120 + 20766785 ACUGCAAUUGCAGUGGCAAUAGCAAUAGCAACCGCGUCGUUUCCGCUGCUCUUGGCCAAAAAAGGCAUUCGACAAUUAUUGCUAUUGAGUGUGAGUGUUAGAGGAAAGGAAAGGAAAGGC ..(((.(((((.((....)).))))).))).((...((.(((((..(.((((..(((......)))....((((....(..(........)..).)))))))).)..))))).))..)). ( -33.00) >DroSec_CAF1 8782 119 + 1 ACUGCAAUUGCAGUGGCAACAGCAAUAGCAACCGCGUCGUUUCCGCUGCUCUUGGCCAAAAAAGGCAUUCGACAAUUAUUGCUAUUGAGUGUGAGUGUGAGUGG-GAGGAAAGGAAAGGC ..(((.(((((..((....))))))).))).((...((.(((((.(..(((...(((......)))((((.(((.(((.......))).)))))))..)))..)-..))))).))..)). ( -37.20) >DroSim_CAF1 14713 120 + 1 ACUGCAAUUGCAGUGGCAACAGCAAUAGCAACCGCGUCGUUUCCGCUGCUCUUGGCCAAAAAAGGCAUUCGACAAUUAUUGCUAUUGAGUGUGAGUGUGAGUGGAGAGGAAAGGAAAGGC ..(((.(((((..((....))))))).))).((...((.(((((.(..(((...(((......)))((((.(((.(((.......))).)))))))..)))..)...))))).))..)). ( -37.50) >DroYak_CAF1 20465 90 + 1 ACUGCAAUUGCAGUGGCA------------AUUGCAGCGUUUCCGCUGCUUUUGGCCAAAAAAGGCAUUCGACAAUUAUUGUUAUUGAAUGUGA------------------GGAAAGGC ..(((((((((....)))------------))))))((.(((((....(((((......)))))((((((((.(((....))).))))))))..------------------))))).)) ( -32.00) >consensus ACUGCAAUUGCAGUGGCAACAGCAAUAGCAACCGCGUCGUUUCCGCUGCUCUUGGCCAAAAAAGGCAUUCGACAAUUAUUGCUAUUGAGUGUGAGUGUGAGUGG_GAGGAAAGGAAAGGC (((((....))))).(((....(((((((((..(((.((....)).))).....(((......)))............)))))))))..)))............................ (-21.24 = -23.05 + 1.81)

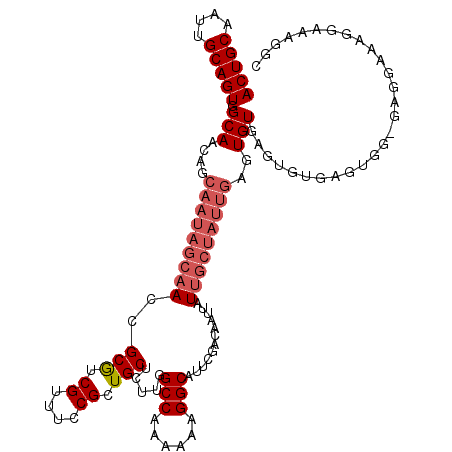

| Location | 17,655,223 – 17,655,343 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.31 |
| Mean single sequence MFE | -30.62 |
| Consensus MFE | -23.23 |
| Energy contribution | -25.35 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17655223 120 - 20766785 GCCUUUCCUUUCCUUUCCUCUAACACUCACACUCAAUAGCAAUAAUUGUCGAAUGCCUUUUUUGGCCAAGAGCAGCGGAAACGACGCGGUUGCUAUUGCUAUUGCCACUGCAAUUGCAGU .................................(((((((((((..........(((......)))(((..((..((....))..))..))).)))))))))))..(((((....))))) ( -31.00) >DroSec_CAF1 8782 119 - 1 GCCUUUCCUUUCCUC-CCACUCACACUCACACUCAAUAGCAAUAAUUGUCGAAUGCCUUUUUUGGCCAAGAGCAGCGGAAACGACGCGGUUGCUAUUGCUGUUGCCACUGCAAUUGCAGU ...............-.................(((((((((((..........(((......)))(((..((..((....))..))..))).)))))))))))..(((((....))))) ( -31.00) >DroSim_CAF1 14713 120 - 1 GCCUUUCCUUUCCUCUCCACUCACACUCACACUCAAUAGCAAUAAUUGUCGAAUGCCUUUUUUGGCCAAGAGCAGCGGAAACGACGCGGUUGCUAUUGCUGUUGCCACUGCAAUUGCAGU .................................(((((((((((..........(((......)))(((..((..((....))..))..))).)))))))))))..(((((....))))) ( -31.00) >DroYak_CAF1 20465 90 - 1 GCCUUUCC------------------UCACAUUCAAUAACAAUAAUUGUCGAAUGCCUUUUUUGGCCAAAAGCAGCGGAAACGCUGCAAU------------UGCCACUGCAAUUGCAGU (((.....------------------...(((((.((((......)))).)))))........))).....((((((....))))))...------------....(((((....))))) ( -29.49) >consensus GCCUUUCCUUUCCUC_CCACUCACACUCACACUCAAUAGCAAUAAUUGUCGAAUGCCUUUUUUGGCCAAGAGCAGCGGAAACGACGCGGUUGCUAUUGCUGUUGCCACUGCAAUUGCAGU .................................(((((((((((..........(((......))).....((..((....))..))......)))))))))))..(((((....))))) (-23.23 = -25.35 + 2.12)



| Location | 17,655,303 – 17,655,413 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 87.10 |
| Mean single sequence MFE | -27.32 |
| Consensus MFE | -21.23 |
| Energy contribution | -21.60 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.557996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17655303 110 + 20766785 GCUAUUGAGUGUGAGUGUUAGAGGAAAGGAAAGGAAAGGCAACAAAUGCAAUACACUGCACUUAAAUUGCGCCCACUAAUAACCCAGUCGAUGGGUAAAAGGGACAGCGG ....((((((((.(((((....................(((.....)))...)))))))))))))..(((.(((.......(((((.....)))))....)))...))). ( -26.40) >DroSec_CAF1 8862 109 + 1 GCUAUUGAGUGUGAGUGUGAGUGG-GAGGAAAGGAAAGGCAACAAGUGCAAUACACUGCACUUAAAUUGCGCCCACUAAUAACCCAGUCGAUGGGUAAAAGGGACAGCGG .(((((((.((.(..(((.(((((-(.(.((......(....)(((((((......)))))))...)).).)))))).)))..))).)))))))................ ( -33.50) >DroSim_CAF1 14793 110 + 1 GCUAUUGAGUGUGAGUGUGAGUGGAGAGGAAAGGAAAGGCAACAAGUGCAAUACACUGCACUUAAAUUGCGCCCACUAAUAACCCAGUCGAUGGGUAAAAGGGACAGCGG .(((((((.((.(..(((.(((((.(.(.((......(....)(((((((......)))))))...)).).)))))).)))..))).)))))))................ ( -29.70) >DroYak_CAF1 20533 92 + 1 GUUAUUGAAUGUGA------------------GGAAAGGCAACAAGUGCAAUGCACUGCACUUAAAUUGCGCCCACUUAUAAUCCAGUCGAUGAGUAAAAGGGACAGCAG ..............------------------.....(....)(((((((......)))))))....(((.(((..((((.(((.....)))..))))..)))...))). ( -19.70) >consensus GCUAUUGAGUGUGAGUGUGAGUGG_GAGGAAAGGAAAGGCAACAAGUGCAAUACACUGCACUUAAAUUGCGCCCACUAAUAACCCAGUCGAUGGGUAAAAGGGACAGCGG ....((((((((.........................(....).((((.....))))))))))))..(((.(((.......(((((.....)))))....)))...))). (-21.23 = -21.60 + 0.37)

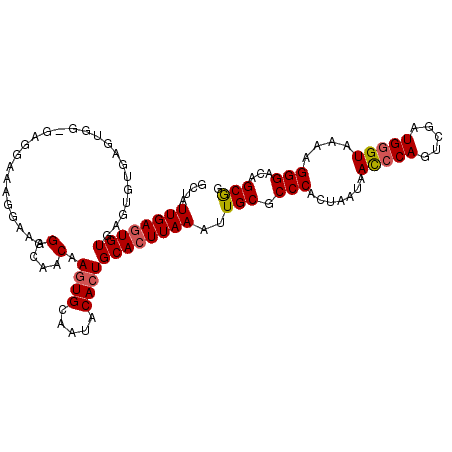
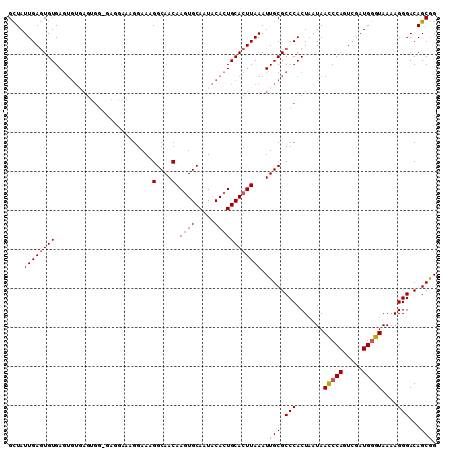
| Location | 17,655,303 – 17,655,413 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 87.10 |
| Mean single sequence MFE | -25.90 |
| Consensus MFE | -24.25 |
| Energy contribution | -24.12 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.29 |
| SVM RNA-class probability | 0.991872 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17655303 110 - 20766785 CCGCUGUCCCUUUUACCCAUCGACUGGGUUAUUAGUGGGCGCAAUUUAAGUGCAGUGUAUUGCAUUUGUUGCCUUUCCUUUCCUUUCCUCUAACACUCACACUCAAUAGC ..(((((((((...(((((.....)))))....)).))))(((((..((((((((....)))))))))))))...................................))) ( -26.00) >DroSec_CAF1 8862 109 - 1 CCGCUGUCCCUUUUACCCAUCGACUGGGUUAUUAGUGGGCGCAAUUUAAGUGCAGUGUAUUGCACUUGUUGCCUUUCCUUUCCUC-CCACUCACACUCACACUCAAUAGC ..(((((((((...(((((.....)))))....)).))))(((((..((((((((....))))))))))))).............-.....................))) ( -28.30) >DroSim_CAF1 14793 110 - 1 CCGCUGUCCCUUUUACCCAUCGACUGGGUUAUUAGUGGGCGCAAUUUAAGUGCAGUGUAUUGCACUUGUUGCCUUUCCUUUCCUCUCCACUCACACUCACACUCAAUAGC ..(((((((((...(((((.....)))))....)).))))(((((..((((((((....)))))))))))))...................................))) ( -28.30) >DroYak_CAF1 20533 92 - 1 CUGCUGUCCCUUUUACUCAUCGACUGGAUUAUAAGUGGGCGCAAUUUAAGUGCAGUGCAUUGCACUUGUUGCCUUUCC------------------UCACAUUCAAUAAC .....(((((((.((.((........)).)).))).))))(((((..((((((((....)))))))))))))......------------------.............. ( -21.00) >consensus CCGCUGUCCCUUUUACCCAUCGACUGGGUUAUUAGUGGGCGCAAUUUAAGUGCAGUGUAUUGCACUUGUUGCCUUUCCUUUCCUC_CCACUCACACUCACACUCAAUAGC .....((((((...(((((.....)))))....)).))))(((((..((((((((....)))))))))))))...................................... (-24.25 = -24.12 + -0.12)

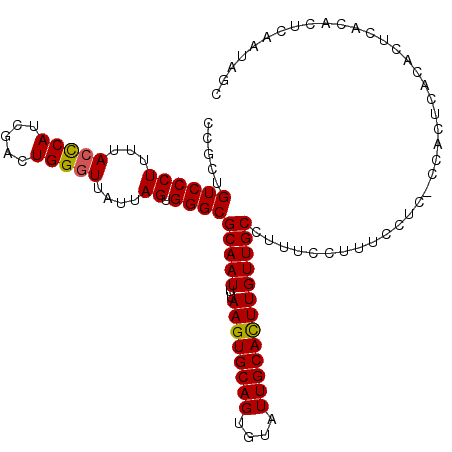
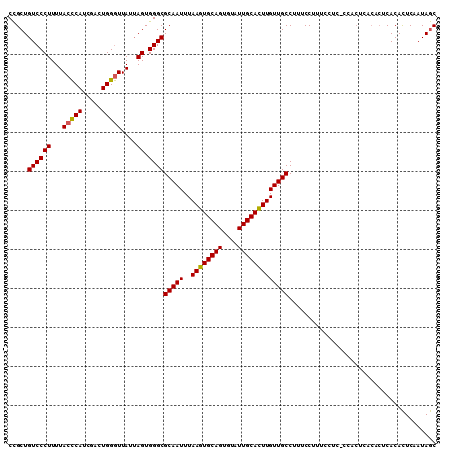
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:56 2006