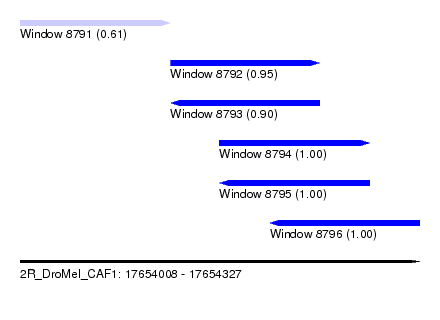
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,654,008 – 17,654,327 |
| Length | 319 |
| Max. P | 0.999692 |

| Location | 17,654,008 – 17,654,128 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.75 |
| Mean single sequence MFE | -23.98 |
| Consensus MFE | -19.22 |
| Energy contribution | -19.10 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.613721 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17654008 120 + 20766785 CCAUGAAAUGAAAACUUUUCCCUCAAAUAUUCAUUAGUUAGUUCCAAGCUCAUUUAAGCAUAGCUUGGGGAAAUAAAAUUUCCAGUUUUGUUAUUUAAAUAGCUGAAUACAUUUCGAAGA ...(((((((((((((...........................(((((((...........)))))))((((((...))))))))))))(((((....)))))......))))))).... ( -23.50) >DroSec_CAF1 7645 120 + 1 CCAUGAAAUGAAAACUUUUCCCCCAAAUAUUCAUUAGUUAGUUCCAAGCUCAUUUGAGCAUAGCUUGGGGAAAUAAAAUUUCCAGUUUUGUUAUUUAAAUGGCUGAAUACAUUUCGAAGA ...(((((((......(((((((............(((((.......((((....)))).))))).)))))))...........((((.(((((....))))).)))).))))))).... ( -23.52) >DroSim_CAF1 13556 116 + 1 CCAUGAAAUGAAAACUUUUCCCCCAAAUAUUCAUUA----GUUCCAAGCUCAUUUGAGCAUAGCUUGGGGAAAUAAAAUUUCCAGUUUUGUUAUUUAAAUAGCUGAAUACAUUUCGAAGA ...(((((((......(((((((............(----(((....((((....))))..)))).)))))))...........((((.(((((....))))).)))).))))))).... ( -23.82) >DroEre_CAF1 7823 119 + 1 CGAUGAUAUGAAAAC-UUUUCCCCAAAUAUUCAUUAGUUAACUCCAAGCUCAUUUGGGCAUGCCUUAGGGAAAUAAAAUGUUCAGUUUUGUUAUUUAAAUAGCUGAAUACAUUUCGAAGA ...(((.(((.....-.((((((............((....))....((((....))))........)))))).....(((((((((.............)))))))))))).))).... ( -21.22) >DroYak_CAF1 19229 120 + 1 CCAUGAAAUGAAAACCUUUUCCCCAAAUAUUCAUUAGUUAACUCCAGGCUCAUUUGGGUAUGCCUUGGGGAAAUAAAAUGAUCAGUUUUGUUAUUUAAAUAGCUGAAUACAUUUCGGAGA ...(((((((.......((((((((((((((((..((((.......))))....)))))))...)))))))))........((((((.............))))))...))))))).... ( -27.82) >consensus CCAUGAAAUGAAAACUUUUCCCCCAAAUAUUCAUUAGUUAGUUCCAAGCUCAUUUGAGCAUAGCUUGGGGAAAUAAAAUUUCCAGUUUUGUUAUUUAAAUAGCUGAAUACAUUUCGAAGA ...(((((((..........((((((.((.....))...........((((....)))).....))))))..............((((.(((((....))))).)))).))))))).... (-19.22 = -19.10 + -0.12)


| Location | 17,654,128 – 17,654,247 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.99 |
| Mean single sequence MFE | -21.40 |
| Consensus MFE | -17.98 |
| Energy contribution | -17.94 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952716 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17654128 119 + 20766785 ACUGAA-AGGCAGAAGCACAAAAACCAUUCGAAAAUCAUCAACAAAUUAAUGUCAAAAUAAUAACAAAUUAACUGCCUUGGCUGCCGUGGCUGCCACAAGAAAUAUGACAGUUUACAAAC ((((.(-((((((.................((......)).(((......)))...................)))))))(((.((....)).))).............))))........ ( -21.50) >DroSec_CAF1 7765 120 + 1 ACUGAACUGGCAGAAGCACAAAAACCAUUCGAAAAUCAUCAACAAAUUAAUGUCAAAAUAAUAACAAAUUAACUGCAUCGGCUGCCGUGGCUGCCACAAGAAAUAUGACAGUUUACAAAC ..(((((((((((.................((......)).(((......)))...................))))...(((.((....)).))).............)))))))..... ( -20.60) >DroSim_CAF1 13672 120 + 1 ACUGAACUGGCAGAAGCACAAAAACCAUUCGAAAAUCAUCAACAAAUUAAUGUCAAAAUAAUAACAAAUUAACUGCCUCGGCUGCCGUGGCUGCCACAAGAAAUAUGACAGUUUACAAAC ..(((((((((((.................((......)).(((......)))...................)))))..(((.((....)).)))..............))))))..... ( -20.00) >DroEre_CAF1 7942 117 + 1 ACUGAAC-GGCAGAAGC--AGAAACCAUUCGAAAAUCAUCAACGAAUUAAUGUCAAAAUAAUAACAAAUUAACUGCCUCGGCUGCCGUGGCUGCCACAAGAAAUAUGACAGUUUACAAAC ((((...-(((((....--.((....(((((...........))))).....))....((((.....)))).)))))..(((.((....)).))).............))))........ ( -22.60) >DroYak_CAF1 19349 117 + 1 ACUGAAC-GGCAGAAGC--AAAAACCAUUCGAAAAUCAUCAACGAAUAAAUGUCAAAAUAAUAACAAAUUAACUGCCUUGGCUGCCGUGGCUGCCACAAGAAAUAUGACAGUUUACAAAC ((((...-(((((....--.......(((((...........)))))...........((((.....)))).))))).((((.((....)).))))............))))........ ( -22.30) >consensus ACUGAAC_GGCAGAAGCACAAAAACCAUUCGAAAAUCAUCAACAAAUUAAUGUCAAAAUAAUAACAAAUUAACUGCCUCGGCUGCCGUGGCUGCCACAAGAAAUAUGACAGUUUACAAAC ((((....(((((.................((......)).(((......)))...................)))))..(((.((....)).))).............))))........ (-17.98 = -17.94 + -0.04)


| Location | 17,654,128 – 17,654,247 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.99 |
| Mean single sequence MFE | -29.88 |
| Consensus MFE | -24.48 |
| Energy contribution | -24.68 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904954 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17654128 119 - 20766785 GUUUGUAAACUGUCAUAUUUCUUGUGGCAGCCACGGCAGCCAAGGCAGUUAAUUUGUUAUUAUUUUGACAUUAAUUUGUUGAUGAUUUUCGAAUGGUUUUUGUGCUUCUGCCU-UUCAGU (.((((...((((((((.....)))))))).....)))).)(((((((...(((((..((((((..((((......))))))))))...)))))(((......))).))))))-)..... ( -27.70) >DroSec_CAF1 7765 120 - 1 GUUUGUAAACUGUCAUAUUUCUUGUGGCAGCCACGGCAGCCGAUGCAGUUAAUUUGUUAUUAUUUUGACAUUAAUUUGUUGAUGAUUUUCGAAUGGUUUUUGUGCUUCUGCCAGUUCAGU .........((((((((.....))))))))....(((((..(.(((((...(((((..((((((..((((......))))))))))...))))).....))))))..)))))........ ( -27.90) >DroSim_CAF1 13672 120 - 1 GUUUGUAAACUGUCAUAUUUCUUGUGGCAGCCACGGCAGCCGAGGCAGUUAAUUUGUUAUUAUUUUGACAUUAAUUUGUUGAUGAUUUUCGAAUGGUUUUUGUGCUUCUGCCAGUUCAGU .........((((((((.....))))))))....(((((..(((((((...(((((..((((((..((((......))))))))))...))))).....)))).))))))))........ ( -28.50) >DroEre_CAF1 7942 117 - 1 GUUUGUAAACUGUCAUAUUUCUUGUGGCAGCCACGGCAGCCGAGGCAGUUAAUUUGUUAUUAUUUUGACAUUAAUUCGUUGAUGAUUUUCGAAUGGUUUCU--GCUUCUGCC-GUUCAGU .........((((((((.....))))))))..((((((...(((((((...(((((..((((((..(((........)))))))))...))))).....))--)))))))))-))..... ( -33.70) >DroYak_CAF1 19349 117 - 1 GUUUGUAAACUGUCAUAUUUCUUGUGGCAGCCACGGCAGCCAAGGCAGUUAAUUUGUUAUUAUUUUGACAUUUAUUCGUUGAUGAUUUUCGAAUGGUUUUU--GCUUCUGCC-GUUCAGU .........((((((((.....))))))))..(((((((...((((((((((..((....))..)))))..(((((((...........)))))))....)--)))))))))-))..... ( -31.60) >consensus GUUUGUAAACUGUCAUAUUUCUUGUGGCAGCCACGGCAGCCGAGGCAGUUAAUUUGUUAUUAUUUUGACAUUAAUUUGUUGAUGAUUUUCGAAUGGUUUUUGUGCUUCUGCC_GUUCAGU .........((((((((.....))))))))....(((((...((((.(((((..((....))..))))).........((((......))))...........)))))))))........ (-24.48 = -24.68 + 0.20)

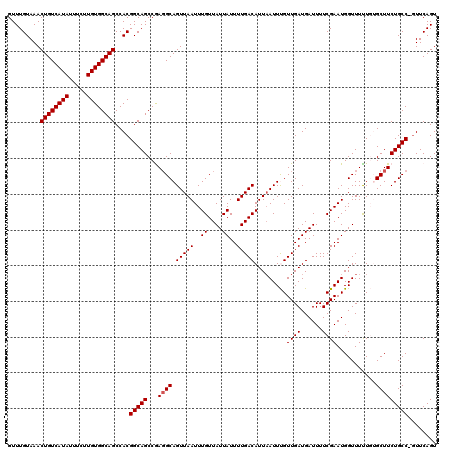
| Location | 17,654,167 – 17,654,287 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -25.64 |
| Consensus MFE | -24.60 |
| Energy contribution | -25.00 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.90 |
| SVM RNA-class probability | 0.999692 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17654167 120 + 20766785 AACAAAUUAAUGUCAAAAUAAUAACAAAUUAACUGCCUUGGCUGCCGUGGCUGCCACAAGAAAUAUGACAGUUUACAAACGGUAAAAAAUGUGCCCCACAGCAGCGGCAAUAAACAUUAA ......(((((((.....((((.....))))..((((...(((((.((((..((..(((....).))(((.(((((.....)))))...))))).)))).)))))))))....))))))) ( -26.40) >DroSec_CAF1 7805 120 + 1 AACAAAUUAAUGUCAAAAUAAUAACAAAUUAACUGCAUCGGCUGCCGUGGCUGCCACAAGAAAUAUGACAGUUUACAAACGGUAAAAAAUGUGCCCCACAGCAGCGGCAAUAAACAUUAA ......(((((((.....((((.....))))..(((....(((((.((((..((..(((....).))(((.(((((.....)))))...))))).)))).))))).)))....))))))) ( -24.10) >DroSim_CAF1 13712 120 + 1 AACAAAUUAAUGUCAAAAUAAUAACAAAUUAACUGCCUCGGCUGCCGUGGCUGCCACAAGAAAUAUGACAGUUUACAAACGGUAAAAAAUGUGCCCCACAGCAGCGGCAAUAAACAUUAA ......(((((((.....((((.....))))..((((...(((((.((((..((..(((....).))(((.(((((.....)))))...))))).)))).)))))))))....))))))) ( -26.40) >DroEre_CAF1 7979 120 + 1 AACGAAUUAAUGUCAAAAUAAUAACAAAUUAACUGCCUCGGCUGCCGUGGCUGCCACAAGAAAUAUGACAGUUUACAAACGGUAAAAAAUGUGCCCCACAGCAGCGGCAAUAAACAUUAA ......(((((((.....((((.....))))..((((...(((((.((((..((..(((....).))(((.(((((.....)))))...))))).)))).)))))))))....))))))) ( -26.40) >DroYak_CAF1 19386 120 + 1 AACGAAUAAAUGUCAAAAUAAUAACAAAUUAACUGCCUUGGCUGCCGUGGCUGCCACAAGAAAUAUGACAGUUUACAAACGGUAAAAAAUGUGCCCCACAGCAGCGGCAAUAAACAUUAA ........(((((.....((((.....))))..((((...(((((.((((..((..(((....).))(((.(((((.....)))))...))))).)))).)))))))))....))))).. ( -24.90) >consensus AACAAAUUAAUGUCAAAAUAAUAACAAAUUAACUGCCUCGGCUGCCGUGGCUGCCACAAGAAAUAUGACAGUUUACAAACGGUAAAAAAUGUGCCCCACAGCAGCGGCAAUAAACAUUAA ......(((((((.....((((.....))))..((((...(((((.((((..((..((.......))(((.(((((.....)))))...))))).)))).)))))))))....))))))) (-24.60 = -25.00 + 0.40)

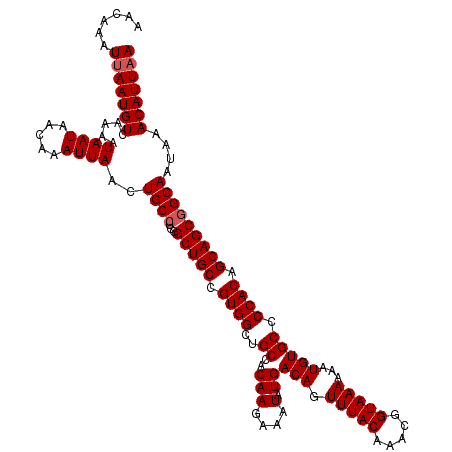
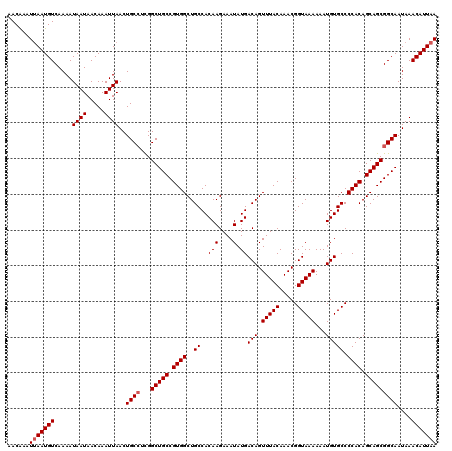
| Location | 17,654,167 – 17,654,287 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -39.32 |
| Consensus MFE | -38.57 |
| Energy contribution | -38.97 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.69 |
| Structure conservation index | 0.98 |
| SVM decision value | 3.74 |
| SVM RNA-class probability | 0.999572 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17654167 120 - 20766785 UUAAUGUUUAUUGCCGCUGCUGUGGGGCACAUUUUUUACCGUUUGUAAACUGUCAUAUUUCUUGUGGCAGCCACGGCAGCCAAGGCAGUUAAUUUGUUAUUAUUUUGACAUUAAUUUGUU ((((((((.((((((((((((((((.........(((((.....)))))((((((((.....))))))))))))))))))...)))))).................))))))))...... ( -40.17) >DroSec_CAF1 7805 120 - 1 UUAAUGUUUAUUGCCGCUGCUGUGGGGCACAUUUUUUACCGUUUGUAAACUGUCAUAUUUCUUGUGGCAGCCACGGCAGCCGAUGCAGUUAAUUUGUUAUUAUUUUGACAUUAAUUUGUU ((((((((.(((((.((((((((((.........(((((.....)))))((((((((.....))))))))))))))))))....))))).................))))))))...... ( -36.77) >DroSim_CAF1 13712 120 - 1 UUAAUGUUUAUUGCCGCUGCUGUGGGGCACAUUUUUUACCGUUUGUAAACUGUCAUAUUUCUUGUGGCAGCCACGGCAGCCGAGGCAGUUAAUUUGUUAUUAUUUUGACAUUAAUUUGUU ((((((((.((((((((((((((((.........(((((.....)))))((((((((.....))))))))))))))))))...)))))).................))))))))...... ( -40.17) >DroEre_CAF1 7979 120 - 1 UUAAUGUUUAUUGCCGCUGCUGUGGGGCACAUUUUUUACCGUUUGUAAACUGUCAUAUUUCUUGUGGCAGCCACGGCAGCCGAGGCAGUUAAUUUGUUAUUAUUUUGACAUUAAUUCGUU ((((((((.((((((((((((((((.........(((((.....)))))((((((((.....))))))))))))))))))...)))))).................))))))))...... ( -40.17) >DroYak_CAF1 19386 120 - 1 UUAAUGUUUAUUGCCGCUGCUGUGGGGCACAUUUUUUACCGUUUGUAAACUGUCAUAUUUCUUGUGGCAGCCACGGCAGCCAAGGCAGUUAAUUUGUUAUUAUUUUGACAUUUAUUCGUU .....(((.((((((((((((((((.........(((((.....)))))((((((((.....))))))))))))))))))...)))))).))).(((((......))))).......... ( -39.30) >consensus UUAAUGUUUAUUGCCGCUGCUGUGGGGCACAUUUUUUACCGUUUGUAAACUGUCAUAUUUCUUGUGGCAGCCACGGCAGCCGAGGCAGUUAAUUUGUUAUUAUUUUGACAUUAAUUUGUU ((((((((.((((((((((((((((.........(((((.....)))))((((((((.....))))))))))))))))))...)))))).................))))))))...... (-38.57 = -38.97 + 0.40)

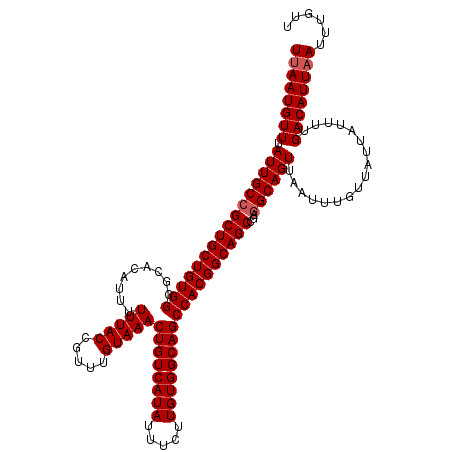

| Location | 17,654,207 – 17,654,327 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.67 |
| Mean single sequence MFE | -37.70 |
| Consensus MFE | -36.90 |
| Energy contribution | -37.10 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.95 |
| SVM RNA-class probability | 0.997857 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17654207 120 - 20766785 AGUUGACGCCUUGAAAAUUGUCAACGUCAUCUAAGCCAAGUUAAUGUUUAUUGCCGCUGCUGUGGGGCACAUUUUUUACCGUUUGUAAACUGUCAUAUUUCUUGUGGCAGCCACGGCAGC ((.(((((..((((......))))))))).)).......................((((((((((.........(((((.....)))))((((((((.....)))))))))))))))))) ( -36.50) >DroSec_CAF1 7845 120 - 1 AGUUGACGCCUUGGAAAUUGUCAACGUCAUCUAAGCCAAGUUAAUGUUUAUUGCCGCUGCUGUGGGGCACAUUUUUUACCGUUUGUAAACUGUCAUAUUUCUUGUGGCAGCCACGGCAGC .((((((..((((((...((....))...))))))....))))))..........((((((((((.........(((((.....)))))((((((((.....)))))))))))))))))) ( -38.00) >DroSim_CAF1 13752 120 - 1 AGUUGACGCCUUGGAAAUUGUCAACGUCAUCUAAGCCAAGUUAAUGUUUAUUGCCGCUGCUGUGGGGCACAUUUUUUACCGUUUGUAAACUGUCAUAUUUCUUGUGGCAGCCACGGCAGC .((((((..((((((...((....))...))))))....))))))..........((((((((((.........(((((.....)))))((((((((.....)))))))))))))))))) ( -38.00) >DroEre_CAF1 8019 120 - 1 AGUUGACGCCUUGGAAAUUGUCAACGUCAUCUAAGCCAAGUUAAUGUUUAUUGCCGCUGCUGUGGGGCACAUUUUUUACCGUUUGUAAACUGUCAUAUUUCUUGUGGCAGCCACGGCAGC .((((((..((((((...((....))...))))))....))))))..........((((((((((.........(((((.....)))))((((((((.....)))))))))))))))))) ( -38.00) >DroYak_CAF1 19426 120 - 1 AGUUGACGCCUUGGAAAUUGUCAACGUCAUCUAAGCCAAGUUAAUGUUUAUUGCCGCUGCUGUGGGGCACAUUUUUUACCGUUUGUAAACUGUCAUAUUUCUUGUGGCAGCCACGGCAGC .((((((..((((((...((....))...))))))....))))))..........((((((((((.........(((((.....)))))((((((((.....)))))))))))))))))) ( -38.00) >consensus AGUUGACGCCUUGGAAAUUGUCAACGUCAUCUAAGCCAAGUUAAUGUUUAUUGCCGCUGCUGUGGGGCACAUUUUUUACCGUUUGUAAACUGUCAUAUUUCUUGUGGCAGCCACGGCAGC .((((((..((((((...((....))...))))))....))))))..........((((((((((.........(((((.....)))))((((((((.....)))))))))))))))))) (-36.90 = -37.10 + 0.20)

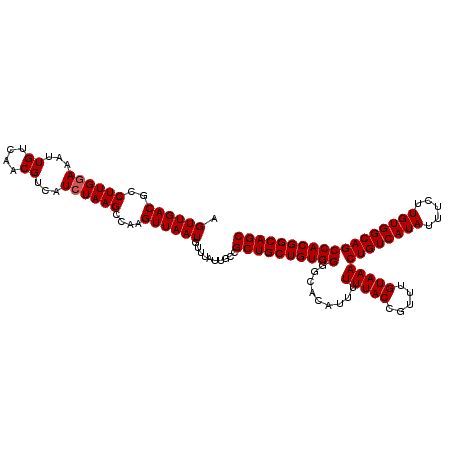
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:46 2006