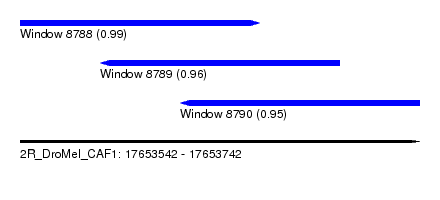
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,653,542 – 17,653,742 |
| Length | 200 |
| Max. P | 0.992460 |

| Location | 17,653,542 – 17,653,662 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.50 |
| Mean single sequence MFE | -31.18 |
| Consensus MFE | -31.16 |
| Energy contribution | -30.68 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.57 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.33 |
| SVM RNA-class probability | 0.992460 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17653542 120 + 20766785 GGAAAAUUGGGCCAACUUAAGCGGCGAUCUGGGGGAAGACUGGCCAAGAACGAUGACUAAGAAAUGUGUUAAUGAUAGCGAGUGGCGUAAUUAGCCAGAAAUAUUUCGUGUUGGCCAAAU .........(((((((......(((.((((......))).).)))...............(((((((((((....))))...((((.......))))...)))))))..))))))).... ( -32.90) >DroSec_CAF1 7175 120 + 1 GGAAAAUUGGGUCAACUUAAGCGGCGAUCUGGGGGAAGACUGGCCAAGAACGAUGACUAAGAAAUGUGUUAAUGAUAGCGAAUGGCGUAAUUAGCCAGAAAUAUUUCGCGUUGGCCAAAA .........(((((((....(((((.((((......))).).)))...............(((((((((((....))))...((((.......))))...)))))))))))))))).... ( -30.90) >DroSim_CAF1 13086 120 + 1 GGAAAAUUGGGUCAACUUAACUGGCGAUCUGGGGGAAGACUGGCCAAGAACAAUGACUAAGAAAUGUGUUAAUGAUAGCGAAUGGCGUAAUUAGCCAGAAAUAUUUCGCGUUGGCCAAAA .........(((((((.....((((.((((......))).).))))....((.((((..(....)..)))).))...(((((((((.......)))).......)))))))))))).... ( -31.31) >DroEre_CAF1 7360 120 + 1 GGAAAAUUGGGUCAACUUGAGCGGCGAUCUGGGGGAAGAGUGGCCAAGAACGAUGACUAAGAAAUGUGUUAAUGAUAGCGAGUGGUGUAAUUAGCCAGAAAUAUUUCUCGUUGGCCAAAA .........(((((((......(((.((((......))).).)))..............((((((((((((....))))...((((.......))))...)))))))).))))))).... ( -28.30) >DroYak_CAF1 18758 120 + 1 GGAAAAUUGGGUCAACUUGAGCGGCGAUCUGGGGGAAGACUGGCCAAGAACGAUGACUAAGAAAUGUGUUAAUGAUAGCGAGUGGCGUAAUUAGCCAGAAACAUUUCUCGUUGGCCAAAA .........(((((((......(((.((((......))).).)))..............((((((((((((....))))...((((.......))))...)))))))).))))))).... ( -32.50) >consensus GGAAAAUUGGGUCAACUUAAGCGGCGAUCUGGGGGAAGACUGGCCAAGAACGAUGACUAAGAAAUGUGUUAAUGAUAGCGAGUGGCGUAAUUAGCCAGAAAUAUUUCGCGUUGGCCAAAA .........(((((((......(((.((((......))).).)))...............(((((((((((....))))...((((.......))))...)))))))..))))))).... (-31.16 = -30.68 + -0.48)

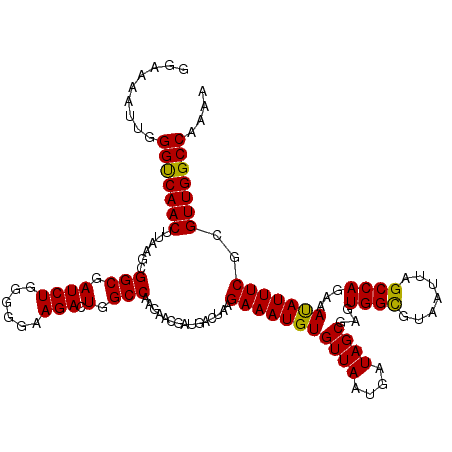
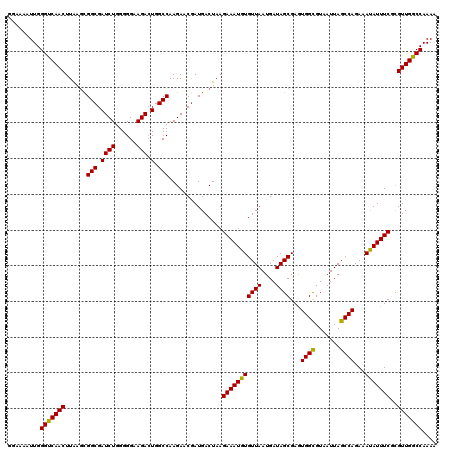
| Location | 17,653,582 – 17,653,702 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -31.58 |
| Consensus MFE | -30.52 |
| Energy contribution | -30.92 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960077 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17653582 120 - 20766785 GCGAGCAUUUUGUGUGGCAUGCGACAGCAAAACGUUUUCGAUUUGGCCAACACGAAAUAUUUCUGGCUAAUUACGCCACUCGCUAUCAUUAACACAUUUCUUAGUCAUCGUUCUUGGCCA (((((.((((((((((((...(((.(((.....))).))).....))).))))))))).....((((.......)))))))))....................((((.......)))).. ( -33.10) >DroSec_CAF1 7215 120 - 1 GCGAGCAUUUUGUGUGGCAUGCGACAGCGGAACGUUUUCGUUUUGGCCAACGCGAAAUAUUUCUGGCUAAUUACGCCAUUCGCUAUCAUUAACACAUUUCUUAGUCAUCGUUCUUGGCCA (((((.((((((((((((..((((.((((...)))).))))....))).))))))))).....((((.......)))))))))....................((((.......)))).. ( -33.20) >DroSim_CAF1 13126 120 - 1 GCGAGCAUUUUGUGUGGCAUGCGACAGCGAAACGUUUUCGUUUUGGCCAACGCGAAAUAUUUCUGGCUAAUUACGCCAUUCGCUAUCAUUAACACAUUUCUUAGUCAUUGUUCUUGGCCA (((((.((((((((((((..((((.((((...)))).))))....))).))))))))).....((((.......)))))))))....................((((.......)))).. ( -33.20) >DroEre_CAF1 7400 120 - 1 GCGAGCAUUUUGUGUGGCAUGCGACAGCAAAACGUUUUCGUUUUGGCCAACGAGAAAUAUUUCUGGCUAAUUACACCACUCGCUAUCAUUAACACAUUUCUUAGUCAUCGUUCUUGGCCA (((((......((((((...((((.(((.....))).)))).(((((((..((((....)))))))))))))))))..)))))....................((((.......)))).. ( -26.40) >DroYak_CAF1 18798 120 - 1 GCGAGCAUUUUGUGUGGCAUGCGACAGCGAAACGUUUUCGUUUUGGCCAACGAGAAAUGUUUCUGGCUAAUUACGCCACUCGCUAUCAUUAACACAUUUCUUAGUCAUCGUUCUUGGCCA (((((((((((.((((((..((((.((((...)))).))))....))).))).))))))....((((.......)))))))))....................((((.......)))).. ( -32.00) >consensus GCGAGCAUUUUGUGUGGCAUGCGACAGCGAAACGUUUUCGUUUUGGCCAACGCGAAAUAUUUCUGGCUAAUUACGCCACUCGCUAUCAUUAACACAUUUCUUAGUCAUCGUUCUUGGCCA (((((.((((((((((((..((((.(((.....))).))))....))).))))))))).....((((.......)))))))))....................((((.......)))).. (-30.52 = -30.92 + 0.40)

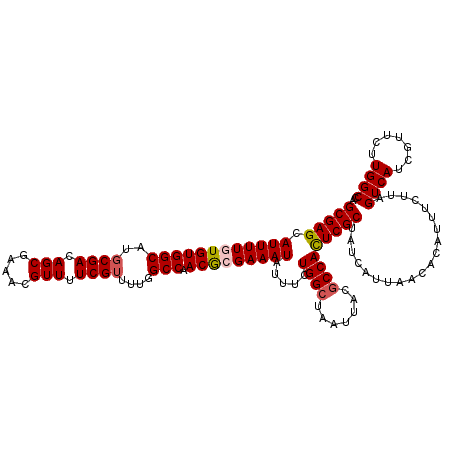
| Location | 17,653,622 – 17,653,742 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.92 |
| Mean single sequence MFE | -35.48 |
| Consensus MFE | -31.82 |
| Energy contribution | -32.34 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946123 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
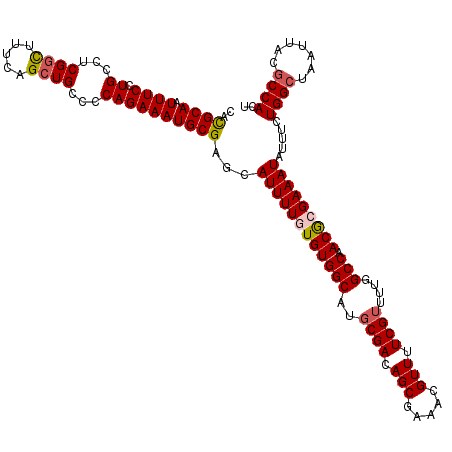
>2R_DroMel_CAF1 17653622 120 - 20766785 CAUGCAAUUUCCUGCUUCGGCUUUCAGCUGCCCCAGAAAUGCGAGCAUUUUGUGUGGCAUGCGACAGCAAAACGUUUUCGAUUUGGCCAACACGAAAUAUUUCUGGCUAAUUACGCCACU ..((((.((((.((...((((.....))))...))))))))))...((((((((((((...(((.(((.....))).))).....))).))))))))).....((((.......)))).. ( -33.40) >DroSec_CAF1 7255 120 - 1 CACGCAAUUUCCUGCCACGGCUUUCAGCUGCCCCAGAAAUGCGAGCAUUUUGUGUGGCAUGCGACAGCGGAACGUUUUCGUUUUGGCCAACGCGAAAUAUUUCUGGCUAAUUACGCCAUU ..((((.((((.((...((((.....))))...))))))))))...((((((((((((..((((.((((...)))).))))....))).))))))))).....((((.......)))).. ( -37.60) >DroSim_CAF1 13166 120 - 1 CACGCAAUUUCCUGCCUCGGCUUUCAGCUGCCCCAGAAAUGCGAGCAUUUUGUGUGGCAUGCGACAGCGAAACGUUUUCGUUUUGGCCAACGCGAAAUAUUUCUGGCUAAUUACGCCAUU ..((((.((((.((...((((.....))))...))))))))))...((((((((((((..((((.((((...)))).))))....))).))))))))).....((((.......)))).. ( -37.60) >DroEre_CAF1 7440 120 - 1 CACGCAAUUUCCUGCCUCGAUUUUCAGCUGCCCCAGAAAUGCGAGCAUUUUGUGUGGCAUGCGACAGCAAAACGUUUUCGUUUUGGCCAACGAGAAAUAUUUCUGGCUAAUUACACCACU ...(((......)))...........((((((.((((((((....)))))).)).)))).)).....(((((((....)))))))((((..((((....))))))))............. ( -28.10) >DroYak_CAF1 18838 120 - 1 CACGCAAUUUCCUGCCGCGGCUUUCAGCUGCCCCAGAAAUGCGAGCAUUUUGUGUGGCAUGCGACAGCGAAACGUUUUCGUUUUGGCCAACGAGAAAUGUUUCUGGCUAAUUACGCCACU ..((((.((((.((..(((((.....)))))..))))))))))((((((((.((((((..((((.((((...)))).))))....))).))).))))))))..((((.......)))).. ( -40.70) >consensus CACGCAAUUUCCUGCCUCGGCUUUCAGCUGCCCCAGAAAUGCGAGCAUUUUGUGUGGCAUGCGACAGCGAAACGUUUUCGUUUUGGCCAACGCGAAAUAUUUCUGGCUAAUUACGCCACU ..((((.((((.((...((((.....))))...))))))))))...((((((((((((..((((.(((.....))).))))....))).))))))))).....((((.......)))).. (-31.82 = -32.34 + 0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:40 2006