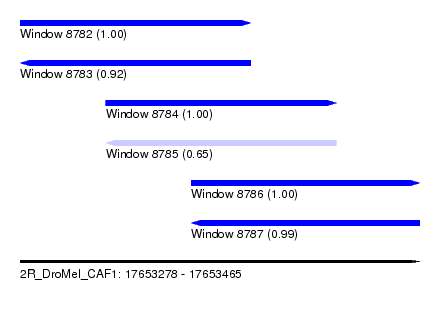
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,653,278 – 17,653,465 |
| Length | 187 |
| Max. P | 0.999811 |

| Location | 17,653,278 – 17,653,386 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.46 |
| Mean single sequence MFE | -43.68 |
| Consensus MFE | -38.40 |
| Energy contribution | -38.68 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.88 |
| SVM decision value | 4.14 |
| SVM RNA-class probability | 0.999811 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17653278 108 + 20766785 CUUGCGUUUUCCGCCGAAUGGCCGCUGGGCAUCGGCAAGUUGCUGAUCCACAAAUUAGUCUCACAUUUAAUUGCAGUGUUCGGCGGCGGGAAAAUGCAGUUG------------CAGUUG .((((((((((((((((...(((....))).)))))..((((((((..(((.((((((........))))))...))).)))))))).)))))))))))...------------...... ( -43.90) >DroSec_CAF1 6908 108 + 1 CUUGCAUUUUCCGCCAAAUGGCCGCUGGGCAUCGGCAAGUUGCUGAUCCACAAAUUAGUCUCACAUUUAAUUGCAGUGCUCGGCGGCGGGAAAAUGCAGUUG------------CAGUUG .(((((((((((..(....)((((((((((((..((((..((.(((..(........)..))))).....)))).)))))))))))).)))))))))))...------------...... ( -42.00) >DroSim_CAF1 12819 108 + 1 CUUGCAUUUUCCGCCGAAUGGCCGCUGGGCAUCGGCAAGUUGGUGAUCCACAAAUUAGUCUCACAUUUAAUUGCAGUGCUCGGCGGCGGGAAAAUGCAGUUG------------CAGUUG .(((((((((((..(....)((((((((((((..((((....((((..(........)..))))......)))).)))))))))))).)))))))))))...------------...... ( -44.30) >DroEre_CAF1 7096 108 + 1 CUUGCAUUUUCCGCCGAAUGGCCGCUGGGCAGCGGCAAGUUGCUGAUCCACAAAUUAGGCUCACAUUUAAUUGCAGUGCUCGGCGGCGGGAAAAUGCAGUUG------------CAGUUG .(((((((((((..(....)(((((((((((...((((..((.(((.((........)).))))).....))))..))))))))))).)))))))))))...------------...... ( -43.20) >DroYak_CAF1 18481 120 + 1 CUUGCAUUUUCCGCCGAAUGGCCGCUGGGCAGCGCCAAGUUGCUGAUCCACAAAUUAGACUCACAUUUAAUUGCAGUGCUUGGCGGCGGGAAAAUGCAGUUGCAGUUGGAGUUGCAGUUG .((((((((((((((.....(((....)))...(((((((.((((.......(((((((......))))))).)))))))))))))).)))))))))))((((((......))))))... ( -45.00) >consensus CUUGCAUUUUCCGCCGAAUGGCCGCUGGGCAUCGGCAAGUUGCUGAUCCACAAAUUAGUCUCACAUUUAAUUGCAGUGCUCGGCGGCGGGAAAAUGCAGUUG____________CAGUUG .(((((((((((((((....(((....)))..))))..((((((((..(((.((((((........))))))...))).)))))))).)))))))))))..................... (-38.40 = -38.68 + 0.28)

| Location | 17,653,278 – 17,653,386 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.46 |
| Mean single sequence MFE | -35.27 |
| Consensus MFE | -28.27 |
| Energy contribution | -29.07 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918133 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17653278 108 - 20766785 CAACUG------------CAACUGCAUUUUCCCGCCGCCGAACACUGCAAUUAAAUGUGAGACUAAUUUGUGGAUCAGCAACUUGCCGAUGCCCAGCGGCCAUUCGGCGGAAAACGCAAG ......------------....(((.((((((.(((((((..(((...........)))........(((.(((((.((.....)).))).))))))))).....))))))))).))).. ( -28.60) >DroSec_CAF1 6908 108 - 1 CAACUG------------CAACUGCAUUUUCCCGCCGCCGAGCACUGCAAUUAAAUGUGAGACUAAUUUGUGGAUCAGCAACUUGCCGAUGCCCAGCGGCCAUUUGGCGGAAAAUGCAAG ......------------....((((((((((.(((((((.((((.((((.....(((((..(........)..))).))..)))).).)))....)))).....))))))))))))).. ( -33.00) >DroSim_CAF1 12819 108 - 1 CAACUG------------CAACUGCAUUUUCCCGCCGCCGAGCACUGCAAUUAAAUGUGAGACUAAUUUGUGGAUCACCAACUUGCCGAUGCCCAGCGGCCAUUCGGCGGAAAAUGCAAG ......------------....((((((((((.(((((((.((((.((((......((((..(........)..))))....)))).).)))....)))).....))))))))))))).. ( -35.60) >DroEre_CAF1 7096 108 - 1 CAACUG------------CAACUGCAUUUUCCCGCCGCCGAGCACUGCAAUUAAAUGUGAGCCUAAUUUGUGGAUCAGCAACUUGCCGCUGCCCAGCGGCCAUUCGGCGGAAAAUGCAAG ......------------....((((((((((.(((((.((...(..(((.................)))..).)).)).....((((((....)))))).....))))))))))))).. ( -37.73) >DroYak_CAF1 18481 120 - 1 CAACUGCAACUCCAACUGCAACUGCAUUUUCCCGCCGCCAAGCACUGCAAUUAAAUGUGAGUCUAAUUUGUGGAUCAGCAACUUGGCGCUGCCCAGCGGCCAUUCGGCGGAAAAUGCAAG ....((((........))))..((((((((((.(((((((((...(((.........(((.((((.....)))))))))).))))))(((((...))))).....))))))))))))).. ( -41.40) >consensus CAACUG____________CAACUGCAUUUUCCCGCCGCCGAGCACUGCAAUUAAAUGUGAGACUAAUUUGUGGAUCAGCAACUUGCCGAUGCCCAGCGGCCAUUCGGCGGAAAAUGCAAG ......................((((((((((.(((((.((...(..(((.................)))..).)).)).....((((........)))).....))))))))))))).. (-28.27 = -29.07 + 0.80)

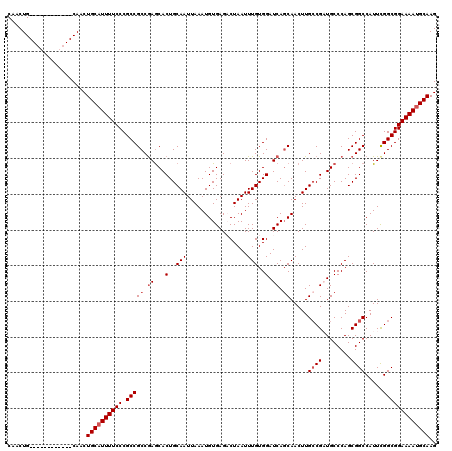
| Location | 17,653,318 – 17,653,426 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.82 |
| Mean single sequence MFE | -35.70 |
| Consensus MFE | -31.40 |
| Energy contribution | -32.00 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.01 |
| SVM RNA-class probability | 0.998120 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17653318 108 + 20766785 UGCUGAUCCACAAAUUAGUCUCACAUUUAAUUGCAGUGUUCGGCGGCGGGAAAAUGCAGUUG------------CAGUUGCAAUUUCGCCUUGCGACUGUCAGCAAGCCGAAAAAAUGCA .((((((......))))))............((((...((((((.(((......))).((((------------(((((((((.......))))))))).))))..))))))....)))) ( -36.30) >DroSec_CAF1 6948 108 + 1 UGCUGAUCCACAAAUUAGUCUCACAUUUAAUUGCAGUGCUCGGCGGCGGGAAAAUGCAGUUG------------CAGUUGCAAUUCCGCCUUGCGACUGUCAGCAAGCCGAAAAAAUGCA .((((((......))))))............((((....((((((((((((...(((((...------------...))))).)))))))((((........))))))))).....)))) ( -34.70) >DroSim_CAF1 12859 108 + 1 UGGUGAUCCACAAAUUAGUCUCACAUUUAAUUGCAGUGCUCGGCGGCGGGAAAAUGCAGUUG------------CAGUUGCAAUUCCGCCUUGCGUCUGUCAGCAAGCCGAAAAAAUGCA ..((((..(........)..)))).......((((....((((((((((((...(((((...------------...))))).)))))))((((........))))))))).....)))) ( -35.80) >DroEre_CAF1 7136 108 + 1 UGCUGAUCCACAAAUUAGGCUCACAUUUAAUUGCAGUGCUCGGCGGCGGGAAAAUGCAGUUG------------CAGUUGCAAUUUCGCCUUGCGACUGUCAGCAAGCCGAAAAAAUGCA ((.(((.((........)).)))))......((((....(((((.(((......))).((((------------(((((((((.......))))))))).))))..))))).....)))) ( -34.70) >DroYak_CAF1 18521 120 + 1 UGCUGAUCCACAAAUUAGACUCACAUUUAAUUGCAGUGCUUGGCGGCGGGAAAAUGCAGUUGCAGUUGGAGUUGCAGUUGCAAUUUCGCCUUGCGACUGUCAGCAAACCGAAAAAAUGCA .((..(..(((.(((((((......)))))))...))).)..))..(((.....(((((((((((.(((((((((....))))))))).).)))))))).)).....))).......... ( -37.00) >consensus UGCUGAUCCACAAAUUAGUCUCACAUUUAAUUGCAGUGCUCGGCGGCGGGAAAAUGCAGUUG____________CAGUUGCAAUUUCGCCUUGCGACUGUCAGCAAGCCGAAAAAAUGCA ..(((((......))))).............((((....((((((((((((...((((..(((((......)))))..)))).)))))))((((........))))))))).....)))) (-31.40 = -32.00 + 0.60)

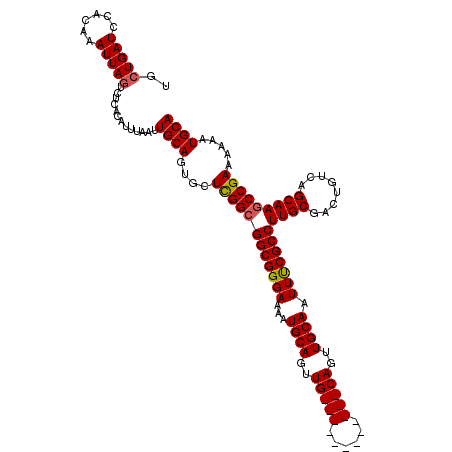
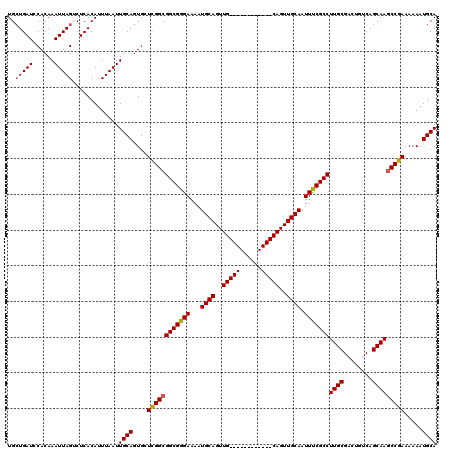
| Location | 17,653,318 – 17,653,426 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.82 |
| Mean single sequence MFE | -31.40 |
| Consensus MFE | -27.00 |
| Energy contribution | -27.04 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.648439 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17653318 108 - 20766785 UGCAUUUUUUCGGCUUGCUGACAGUCGCAAGGCGAAAUUGCAACUG------------CAACUGCAUUUUCCCGCCGCCGAACACUGCAAUUAAAUGUGAGACUAAUUUGUGGAUCAGCA ((((....((((((((((........))))(((((((.((((....------------....)))).))))..)))))))))...))))........(((..(........)..)))... ( -29.50) >DroSec_CAF1 6948 108 - 1 UGCAUUUUUUCGGCUUGCUGACAGUCGCAAGGCGGAAUUGCAACUG------------CAACUGCAUUUUCCCGCCGCCGAGCACUGCAAUUAAAUGUGAGACUAAUUUGUGGAUCAGCA ((((....((((((((((........))))(((((((.((((....------------....))))..)).)))))))))))...))))........(((..(........)..)))... ( -31.50) >DroSim_CAF1 12859 108 - 1 UGCAUUUUUUCGGCUUGCUGACAGACGCAAGGCGGAAUUGCAACUG------------CAACUGCAUUUUCCCGCCGCCGAGCACUGCAAUUAAAUGUGAGACUAAUUUGUGGAUCACCA ((((....((((((((((........))))(((((((.((((....------------....))))..)).)))))))))))...)))).......((((..(........)..)))).. ( -34.00) >DroEre_CAF1 7136 108 - 1 UGCAUUUUUUCGGCUUGCUGACAGUCGCAAGGCGAAAUUGCAACUG------------CAACUGCAUUUUCCCGCCGCCGAGCACUGCAAUUAAAUGUGAGCCUAAUUUGUGGAUCAGCA ((((....((((((((((........))))(((((((.((((....------------....)))).))))..)))))))))...))))........(((.((........)).)))... ( -30.30) >DroYak_CAF1 18521 120 - 1 UGCAUUUUUUCGGUUUGCUGACAGUCGCAAGGCGAAAUUGCAACUGCAACUCCAACUGCAACUGCAUUUUCCCGCCGCCAAGCACUGCAAUUAAAUGUGAGUCUAAUUUGUGGAUCAGCA ...............((((((...(((((((((((((.((((..((((........))))..)))).))))..)))....((.((((((......))).)))))...)))))).)))))) ( -31.70) >consensus UGCAUUUUUUCGGCUUGCUGACAGUCGCAAGGCGAAAUUGCAACUG____________CAACUGCAUUUUCCCGCCGCCGAGCACUGCAAUUAAAUGUGAGACUAAUUUGUGGAUCAGCA ((((....((((((((((........))))(((((((.((((..((((........))))..)))).))))..)))))))))...))))........(((..(........)..)))... (-27.00 = -27.04 + 0.04)

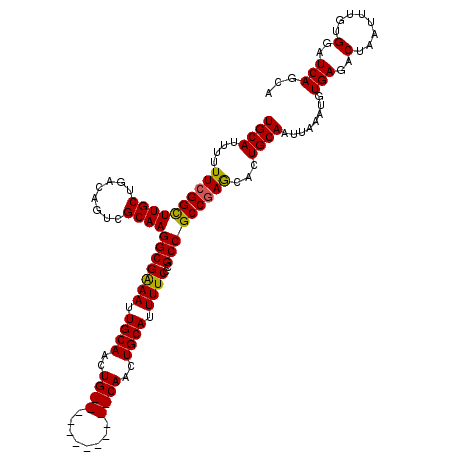
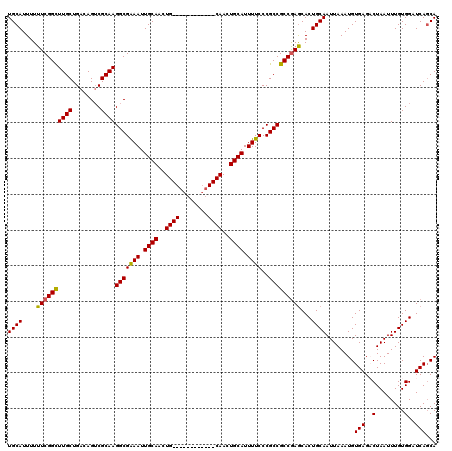
| Location | 17,653,358 – 17,653,465 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.05 |
| Mean single sequence MFE | -36.18 |
| Consensus MFE | -30.88 |
| Energy contribution | -31.48 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.72 |
| SVM RNA-class probability | 0.996584 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17653358 107 + 20766785 CGGCGGCGGGAAAAUGCAGUUG------------CAGUUGCAAUUUCGCCUUGCGACUGUCAGCAAGCCGAAAAAAUGCAGAAAA-AAAUGUUAAUUUCAUUGCGCGGGUAAAAGUCGCC .((((((.......(((.((((------------(((((((((.......))))))))).))))..((.((((....(((.....-...)))...))))...))))).......)))))) ( -34.34) >DroSec_CAF1 6988 107 + 1 CGGCGGCGGGAAAAUGCAGUUG------------CAGUUGCAAUUCCGCCUUGCGACUGUCAGCAAGCCGAAAAAAUGCAGCAAA-AAAUGUUAAUUUCAUUGCGCGGGUAAAAGUCGCC .((((((.......(((.((((------------(((((((((.......))))))))).))))..((.((((....(((.....-...)))...))))...))))).......)))))) ( -34.34) >DroSim_CAF1 12899 107 + 1 CGGCGGCGGGAAAAUGCAGUUG------------CAGUUGCAAUUCCGCCUUGCGUCUGUCAGCAAGCCGAAAAAAUGCAGCAAA-AAAUGUUAAUUUCAUUGCGCGGGUAAAAGUCGCC (((((((((((...(((((...------------...))))).)))))))((((........))))))))..........((((.-((((....))))..))))((((.......)))). ( -32.90) >DroEre_CAF1 7176 107 + 1 CGGCGGCGGGAAAAUGCAGUUG------------CAGUUGCAAUUUCGCCUUGCGACUGUCAGCAAGCCGAAAAAAUGCAGCAAA-AAAUGUUAAUUUCAUUGCGCGGGUAAAAGUCGCC .((((((..((((.(((....)------------))((((((.(((((.(((((........))))).)))))...))))))...-.........)))).((((....))))..)))))) ( -35.70) >DroYak_CAF1 18561 120 + 1 UGGCGGCGGGAAAAUGCAGUUGCAGUUGGAGUUGCAGUUGCAAUUUCGCCUUGCGACUGUCAGCAAACCGAAAAAAUGCAGCAAAAAAAUGUUCAUUUCAUUGCGCGGGUAAAAGUCGCC .((((((((.....(((((((((((.(((((((((....))))))))).).)))))))).)).....)).......(((.((((..((((....))))..))))))).......)))))) ( -43.60) >consensus CGGCGGCGGGAAAAUGCAGUUG____________CAGUUGCAAUUUCGCCUUGCGACUGUCAGCAAGCCGAAAAAAUGCAGCAAA_AAAUGUUAAUUUCAUUGCGCGGGUAAAAGUCGCC (((((((((((...((((..(((((......)))))..)))).)))))))((((........))))))))..........((((..((((....))))..))))((((.......)))). (-30.88 = -31.48 + 0.60)

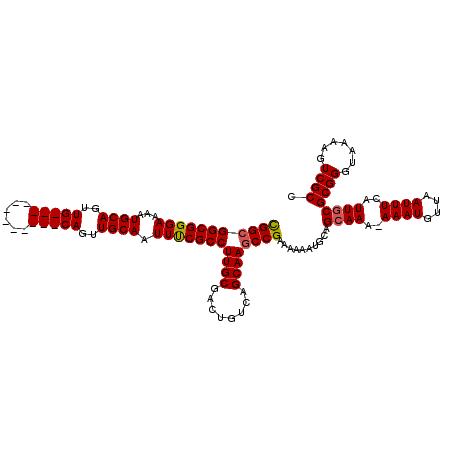
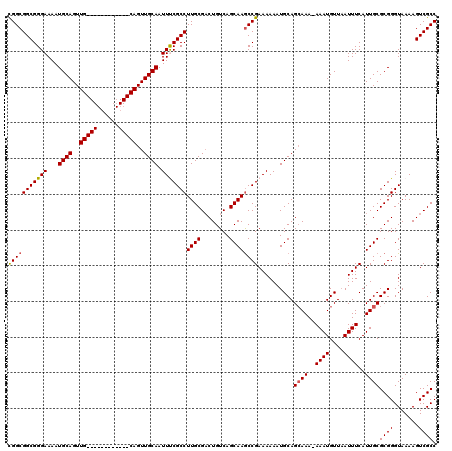
| Location | 17,653,358 – 17,653,465 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.05 |
| Mean single sequence MFE | -32.50 |
| Consensus MFE | -27.68 |
| Energy contribution | -27.88 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987499 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17653358 107 - 20766785 GGCGACUUUUACCCGCGCAAUGAAAUUAACAUUU-UUUUCUGCAUUUUUUCGGCUUGCUGACAGUCGCAAGGCGAAAUUGCAACUG------------CAACUGCAUUUUCCCGCCGCCG ((((..........(((((..((((........)-)))..)))....(((((.(((((........))))).)))))..))...((------------(....)))......)))).... ( -29.60) >DroSec_CAF1 6988 107 - 1 GGCGACUUUUACCCGCGCAAUGAAAUUAACAUUU-UUUGCUGCAUUUUUUCGGCUUGCUGACAGUCGCAAGGCGGAAUUGCAACUG------------CAACUGCAUUUUCCCGCCGCCG (((((((.......((((((.(((((....))))-))))).))......((((....)))).))))....(((((((.((((....------------....))))..)).)))))))). ( -32.20) >DroSim_CAF1 12899 107 - 1 GGCGACUUUUACCCGCGCAAUGAAAUUAACAUUU-UUUGCUGCAUUUUUUCGGCUUGCUGACAGACGCAAGGCGGAAUUGCAACUG------------CAACUGCAUUUUCCCGCCGCCG ((((..........((((((.(((((....))))-)))))((((...(((((.(((((........))))).))))).))))...)------------)....((........)))))). ( -31.40) >DroEre_CAF1 7176 107 - 1 GGCGACUUUUACCCGCGCAAUGAAAUUAACAUUU-UUUGCUGCAUUUUUUCGGCUUGCUGACAGUCGCAAGGCGAAAUUGCAACUG------------CAACUGCAUUUUCCCGCCGCCG ((((..........((((((.(((((....))))-)))))((((...(((((.(((((........))))).))))).))))...)------------)....((........)))))). ( -31.90) >DroYak_CAF1 18561 120 - 1 GGCGACUUUUACCCGCGCAAUGAAAUGAACAUUUUUUUGCUGCAUUUUUUCGGUUUGCUGACAGUCGCAAGGCGAAAUUGCAACUGCAACUCCAACUGCAACUGCAUUUUCCCGCCGCCA (((((((.......((((((.(((((....))))).)))).))......((((....)))).))))....(((((((.((((..((((........))))..)))).))))..)))))). ( -37.40) >consensus GGCGACUUUUACCCGCGCAAUGAAAUUAACAUUU_UUUGCUGCAUUUUUUCGGCUUGCUGACAGUCGCAAGGCGAAAUUGCAACUG____________CAACUGCAUUUUCCCGCCGCCG (((...........((((((..((((....))))..)))).))........(((((((........))))((.((((.((((..((((........))))..)))).)))))))))))). (-27.68 = -27.88 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:37 2006