
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,652,856 – 17,653,148 |
| Length | 292 |
| Max. P | 0.999784 |

| Location | 17,652,856 – 17,652,976 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.61 |
| Mean single sequence MFE | -37.42 |
| Consensus MFE | -34.50 |
| Energy contribution | -34.06 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.92 |
| SVM decision value | 4.07 |
| SVM RNA-class probability | 0.999784 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
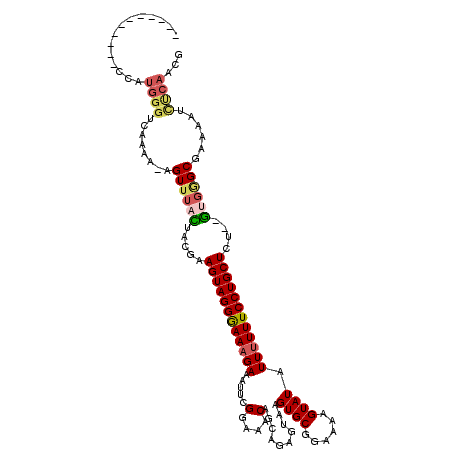
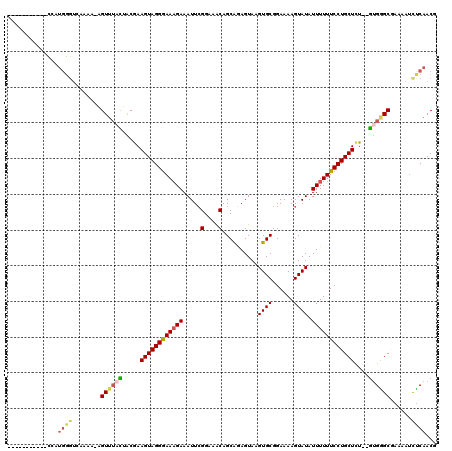
>2R_DroMel_CAF1 17652856 120 + 20766785 UUGCUGGCCACGUUAAUGUGGCCAUCAUAAAAAGCGUGUGAACCCAGUGUUAACGUGUCGAAGCCUAGACAAUAAGCCACACUUGGCGACACUUUUCCCUGGCCUUCGCUUUUCCCAGCU ..((((((((((....)))))))......(((((((.......((((.(..((.(((((...(......).....((((....))))))))).))..)))))....)))))))...))). ( -37.80) >DroSec_CAF1 6470 118 + 1 UUGCUGGCCACGUUAAUGUGGCCACCAUAAAAAGCGUGUGAACCCAGUGCUAACGUGUCGAA--CCAGACAAUAAGCCACACUUGGCGACACUUUUCCCUGGCCUUCGCUUUUCCCAGCU ..((((((((((....)))))))......(((((((.......((((.(..((.((((((..--.....).....((((....))))))))).))..)))))....)))))))...))). ( -36.40) >DroSim_CAF1 12368 118 + 1 UUGCUGGCCACGUUAAUGUGGCCACCAUAAAAAGCGUGUGAACCCAGUGUUAACGUGUCGAA--CCAGACAAUAAGCCACACUUGGCGACACUUUUCCCUGGCCUUCGCUUUUCCCAGCU ..((((((((((....)))))))......(((((((.......((((.(..((.((((((..--.....).....((((....))))))))).))..)))))....)))))))...))). ( -37.40) >DroEre_CAF1 6591 118 + 1 UUGUUGGCCACGUUAAUGUGGCCACCAUAAAAAGCGUGUGAACCAAGUGUUAACGUGUCGAA--CCAGACAAUAAGUCGCACUUGGCGACACUUUUCCCUGGCCAUCGCUUUUCCUCGCU .((.((((((((....)))))))).))..((((((((((...((((((((..((.((((...--...))))....)).))))))))..))))....(....).....))))))....... ( -37.20) >DroYak_CAF1 17947 118 + 1 UUGUUGGCCACGUUAAUGUGGCCACCAUAAAAAGCGUGUGAACCCAGUGUUAACGUGUCGAA--CCAGACAAUAAGCCACACUUGGCGACACUUUUCUCUGGCCAUCGCUUUUCCUCGCC .((.((((((((....)))))))).))..(((((((.(((...((((.(..((.((((((..--.....).....((((....)))))))))))..).)))).))))))))))....... ( -38.30) >consensus UUGCUGGCCACGUUAAUGUGGCCACCAUAAAAAGCGUGUGAACCCAGUGUUAACGUGUCGAA__CCAGACAAUAAGCCACACUUGGCGACACUUUUCCCUGGCCUUCGCUUUUCCCAGCU .((.((((((((....)))))))).))..(((((((.......((((.(..((.((((((.....).........((((....))))))))).))..)))))....)))))))....... (-34.50 = -34.06 + -0.44)

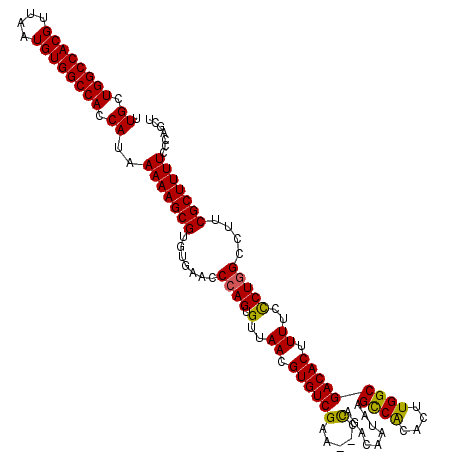
| Location | 17,652,856 – 17,652,976 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.61 |
| Mean single sequence MFE | -42.60 |
| Consensus MFE | -38.34 |
| Energy contribution | -38.78 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.62 |
| SVM RNA-class probability | 0.999460 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17652856 120 - 20766785 AGCUGGGAAAAGCGAAGGCCAGGGAAAAGUGUCGCCAAGUGUGGCUUAUUGUCUAGGCUUCGACACGUUAACACUGGGUUCACACGCUUUUUAUGAUGGCCACAUUAACGUGGCCAGCAA .(((..((((((((.((.(((((..((.((((((..((((.((((.....).))).)))))))))).))..).)))).))....)))))))).....((((((......))))))))).. ( -45.20) >DroSec_CAF1 6470 118 - 1 AGCUGGGAAAAGCGAAGGCCAGGGAAAAGUGUCGCCAAGUGUGGCUUAUUGUCUGG--UUCGACACGUUAGCACUGGGUUCACACGCUUUUUAUGGUGGCCACAUUAACGUGGCCAGCAA ......((((((((.((.((((......(((((((((.....(((.....))))))--..)))))).......)))).))....)))))))).((.(((((((......))))))).)). ( -42.52) >DroSim_CAF1 12368 118 - 1 AGCUGGGAAAAGCGAAGGCCAGGGAAAAGUGUCGCCAAGUGUGGCUUAUUGUCUGG--UUCGACACGUUAACACUGGGUUCACACGCUUUUUAUGGUGGCCACAUUAACGUGGCCAGCAA ......((((((((.((.(((((..((.(((((((((.....(((.....))))))--..)))))).))..).)))).))....)))))))).((.(((((((......))))))).)). ( -42.80) >DroEre_CAF1 6591 118 - 1 AGCGAGGAAAAGCGAUGGCCAGGGAAAAGUGUCGCCAAGUGCGACUUAUUGUCUGG--UUCGACACGUUAACACUUGGUUCACACGCUUUUUAUGGUGGCCACAUUAACGUGGCCAACAA ......((((((((.((((((((.....(((((((((.....(((.....))))))--..)))))).......))))).)))..)))))))).((.(((((((......))))))).)). ( -43.00) >DroYak_CAF1 17947 118 - 1 GGCGAGGAAAAGCGAUGGCCAGAGAAAAGUGUCGCCAAGUGUGGCUUAUUGUCUGG--UUCGACACGUUAACACUGGGUUCACACGCUUUUUAUGGUGGCCACAUUAACGUGGCCAACAA .((........))....((((..(((((((((.(((.((((((((....((((...--...)))).))).))))).)))....))))))))).))))((((((......))))))..... ( -39.50) >consensus AGCUGGGAAAAGCGAAGGCCAGGGAAAAGUGUCGCCAAGUGUGGCUUAUUGUCUGG__UUCGACACGUUAACACUGGGUUCACACGCUUUUUAUGGUGGCCACAUUAACGUGGCCAGCAA ......((((((((.((.((((......((((((........(((.....))).......)))))).......)))).))....)))))))).((.(((((((......))))))).)). (-38.34 = -38.78 + 0.44)

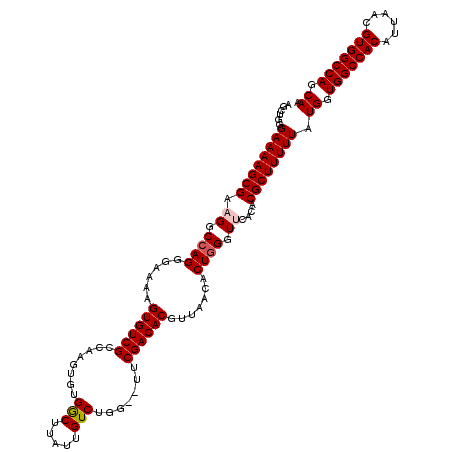

| Location | 17,652,936 – 17,653,055 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.50 |
| Mean single sequence MFE | -30.21 |
| Consensus MFE | -19.99 |
| Energy contribution | -20.82 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.507892 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17652936 119 - 20766785 GAAAUCAUUAGGGGAAAUGGAAAUGGCGAAGAAAAACCGCCUUCAACUUUG-GCCGGUAAAAACGGCUCAUUGGACUUGCAGCUGGGAAAAGCGAAGGCCAGGGAAAAGUGUCGCCAAGU ....(((((......)))))...((((((.......((((((((..(..((-((((.......)))).))..)........(((......)))))))))..))........))))))... ( -35.86) >DroSec_CAF1 6548 107 - 1 GAAAUCAUUAGGGGAAAUGGAAAUGGCGGAGAAAAAUCGCCUUCAACUUUAGG-------------CCCAAUCGAAUUGCAGCUGGGAAAAGCGAAGGCCAGGGAAAAGUGUCGCCAAGU ....(((((......)))))...(((((((......))((((((..((((...-------------((((.............)))).)))).)))))).............)))))... ( -24.62) >DroSim_CAF1 12446 120 - 1 GAAAUCAUUAGGGGAAAUGGAAAUGGCGGAGAAAAACCGCCUUCAACUUUAGGCCGGUAAAAACGGCCCAAUCGACUUGCAGCUGGGAAAAGCGAAGGCCAGGGAAAAGUGUCGCCAAGU ....(((((......)))))...((((((.......((((((((.......(((((.......))))).............(((......)))))))))..))........))))))... ( -35.96) >DroEre_CAF1 6669 96 - 1 GAAAUCAUUAGGGAAAAUGGAAAUGGCGAUGAAAA-CCGCCUUCAACUUU-----------------------GACUUACAGCGAGGAAAAGCGAUGGCCAGGGAAAAGUGUCGCCAAGU ....(((((......)))))...((((((((....-(((((.((..((((-----------------------..(((.....)))..)))).)).)))..))......))))))))... ( -23.70) >DroYak_CAF1 18025 119 - 1 GAAAUCAUUAGGGCGAAUGGAAAUGGUGAAGAAAAGCCGCAUUCACUUUUG-GCCGGUAAAAACGACCCAAUCGACUUACGGCGAGGAAAAGCGAUGGCCAGAGAAAAGUGUCGCCAAGU ...........(((((.((((..((((........))))..)))).(((((-((((((.......)))..((((.(((.(.....)...))))))))))))))).......))))).... ( -30.90) >consensus GAAAUCAUUAGGGGAAAUGGAAAUGGCGAAGAAAAACCGCCUUCAACUUUA_GCCGGUAAAAACG_CCCAAUCGACUUGCAGCUGGGAAAAGCGAAGGCCAGGGAAAAGUGUCGCCAAGU ....(((((......)))))...((((((.......((((((((........((((.......))))..............((........))))))))..))........))))))... (-19.99 = -20.82 + 0.83)


| Location | 17,653,055 – 17,653,148 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.52 |
| Mean single sequence MFE | -25.53 |
| Consensus MFE | -19.88 |
| Energy contribution | -20.72 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.689979 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17653055 93 - 20766785 -------------------------AGUUUACUACGAAGUAGGGAAAGAAAUUCGGAAACAGCAGAGUAAGUGCGGAAAAGUAUAUUUUUUCCUGCUCU--GUGGGCGAAAAUCUCAACG -------------------------......(((((.((((((((((((.....(....)..........((((......)))).)))))))))))).)--))))............... ( -24.30) >DroSec_CAF1 6655 106 - 1 -----------UAAUGGGUCUAAA-AGUUUACUACGAAGUAGGGAAAGAAAUUCGGAAACAGCAGAGUAAGUGCGGAAAAGUAUAUUUUUUCCUGCUCU--GUGAGCGAAAAUUUCAACG -----------...((((......-.((((((...((.(((((((((((.....(....)..........((((......)))).))))))))))))).--))))))......))))... ( -24.22) >DroSim_CAF1 12566 106 - 1 -----------UCAUGGGUCUAAA-AGUUUACUACGAAGUAGGGAAAGAAAUUCGGAAACAGCAGAGUAAGUGCGGAAAAGUAUAUUUUUUCCUGCUCU--GUGAGCGAAAAUCUCAACG -----------...((((......-.((((((...((.(((((((((((.....(....)..........((((......)))).))))))))))))).--))))))......))))... ( -26.52) >DroEre_CAF1 6765 120 - 1 UCUCUGUACUCCCAUGGGUCAACAAAGUUGUUAACAAAGUAGGGAACGAAAUUCGGAAACAGCGCAGUAAGUGCGGAAAAGUAUAUUUUUUCCUGCUCUGUGUGGGCCAAAAACCCAACG ...((((..((((((..((.((((....)))).))...)).)))).((.....))...))))(((((.....(((((((((.....))))))).)).)))))((((.......))))... ( -29.40) >DroYak_CAF1 18144 117 - 1 UCUCUGAUUUUCCGUGGGUAAAAAUUGUGUAUAGCAAAGUAGGAAAAGAAAUU-GGAAGCAGCAAAGUAAGUGCGGAAAAGUAUAUUUUUUCCUGCUUU--AAGGGCUAAAAUCUCAACG .....(((((..((..(.......)..))..((((((((((((((((((..((-(....)))........((((......)))).))))))))))))))--....)))))))))...... ( -23.20) >consensus ___________CCAUGGGUCAAAA_AGUUUACUACGAAGUAGGGAAAGAAAUUCGGAAACAGCAGAGUAAGUGCGGAAAAGUAUAUUUUUUCCUGCUCU__GUGGGCGAAAAUCUCAACG ..............((((........((((((.....((((((((((((.....(....)..........((((......)))).))))))))))))....))))))......))))... (-19.88 = -20.72 + 0.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:32 2006