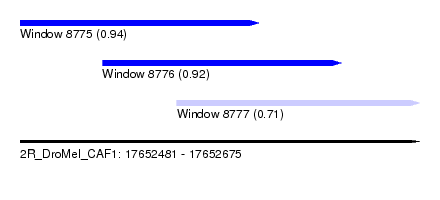
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,652,481 – 17,652,675 |
| Length | 194 |
| Max. P | 0.936198 |

| Location | 17,652,481 – 17,652,597 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.39 |
| Mean single sequence MFE | -25.32 |
| Consensus MFE | -20.70 |
| Energy contribution | -20.30 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936198 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17652481 116 + 20766785 GAUUUGCAUUUAAUUGCGUAUAAUUGGAAAACCCAAUAUGUAUUAUUUUCA----CUUAAUUUUAUUUCACAAAUGCAGCUCGAAAAACGUUUGCUGUCCAGCGAAAGCAAACAAAAGCG ((.((((((((...(((((((...(((.....))))))))))(((......----..)))...........)))))))).)).......(((((((.((....)).)))))))....... ( -23.20) >DroSec_CAF1 6093 116 + 1 GAUUUGCAUUUAAUUGCGUAUAAUUGGAAAACCAAAUAUGUAUUAUUUUCA----CUUAAUUUUAUUUCGCAAAUGCAGCUCGAAAAACGUUUGCUGUCCAGCGAAAGUAAACAAAAACG ..(((((.......(((((((..((((....))))))))))).........----..........((((((....(((((.((.....))...)))))...)))))))))))........ ( -23.80) >DroSim_CAF1 11991 116 + 1 GAUUUGCAUUUAAUUGCGUAUAAUUGGAAAACCAAAUAUGUAUUUUUUUCA----CUUAAUUUUAUUUCGCAAAUGCAGCUCGAAAAACGUUUGCUGUCCAGCGAAAGCAAACAAAAGCG ..(((((.......(((((((..((((....))))))))))).........----..........((((((....(((((.((.....))...)))))...)))))))))))........ ( -25.80) >DroEre_CAF1 6226 116 + 1 GAUUUGCAUUUAAUUGCGUAUAAUUGGAAAACCAAUUAUGUAUUAUUUUCA----CUUAAUUUUAUUUGGCAAAUGCAGCUCGAAAAACGCUUGCUGUCCAGCGAAAGCAAACAGAAGUG ((.(((((((....(((.(((((((((....))))))))).((((......----..))))........)))))))))).))......(((((.((((...((....))..))))))))) ( -29.10) >DroYak_CAF1 17549 120 + 1 GAUUUGCAUUUAAUUGCGUAUAAUUGGAAAACCAAAUAUGUAUUAUUUUCACUCACUUAACUUUAUUUGGCAAAUGCAACUCGAAAAACGUUUGCUGUCCAGCGAAAGCAAACAAAAGCG ((.((((((((...(((((((..((((....)))))))))))..................((......)).)))))))).)).......(((((((.((....)).)))))))....... ( -24.70) >consensus GAUUUGCAUUUAAUUGCGUAUAAUUGGAAAACCAAAUAUGUAUUAUUUUCA____CUUAAUUUUAUUUCGCAAAUGCAGCUCGAAAAACGUUUGCUGUCCAGCGAAAGCAAACAAAAGCG ((.((((((((...(((((((..((((....)))))))))))................((......))...)))))))).))......(((((..(((...((....))..))).))))) (-20.70 = -20.30 + -0.40)

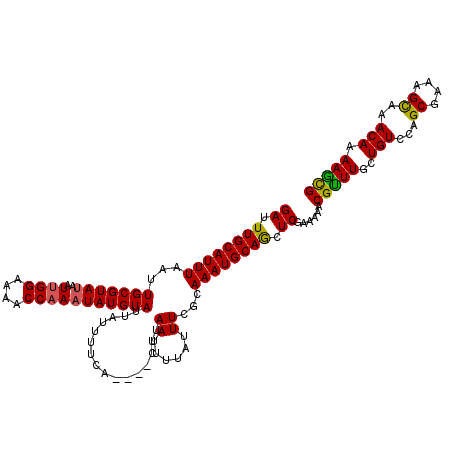
| Location | 17,652,521 – 17,652,637 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.84 |
| Mean single sequence MFE | -26.64 |
| Consensus MFE | -20.60 |
| Energy contribution | -20.92 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915762 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17652521 116 + 20766785 UAUUAUUUUCA----CUUAAUUUUAUUUCACAAAUGCAGCUCGAAAAACGUUUGCUGUCCAGCGAAAGCAAACAAAAGCGCGCAACUGAGGUAACGAGUGCAAUAAAAACCAUUUGUUAA ..(((((..((----(((....((((((((.....(((((.((.....))...)))))...(((...((........)).)))...)))))))).))))).))))).............. ( -22.50) >DroSec_CAF1 6133 116 + 1 UAUUAUUUUCA----CUUAAUUUUAUUUCGCAAAUGCAGCUCGAAAAACGUUUGCUGUCCAGCGAAAGUAAACAAAAACGAGCAACUGAGCUAACGAGUGCAAUAAAAACCAUUUGUUGA .........((----(((...........((....))((((((.....(((((..(((...((....))..))).)))))......))))))...)))))(((((((.....))))))). ( -24.00) >DroSim_CAF1 12031 116 + 1 UAUUUUUUUCA----CUUAAUUUUAUUUCGCAAAUGCAGCUCGAAAAACGUUUGCUGUCCAGCGAAAGCAAACAAAAGCGAGCAACUGAGGUAACGAGUGCAAUAAAAACCAUUUGUUAA ..((((.((((----(((....(((((((((....)).(((((......(((((((.((....)).))))))).....)))))....))))))).))))).)).))))............ ( -27.90) >DroEre_CAF1 6266 116 + 1 UAUUAUUUUCA----CUUAAUUUUAUUUGGCAAAUGCAGCUCGAAAAACGCUUGCUGUCCAGCGAAAGCAAACAGAAGUGAGCGACUGAGGCAAUGAGUGCAAUAAAAAGCAUUUGUUAA ...........----...........(((((((((((..((((.....((((..((.....((....)).......))..))))..))))(((.....)))........))))))))))) ( -30.90) >DroYak_CAF1 17589 120 + 1 UAUUAUUUUCACUCACUUAACUUUAUUUGGCAAAUGCAACUCGAAAAACGUUUGCUGUCCAGCGAAAGCAAACAAAAGCGAGAGACUGAGGUAAUGAGUGCAAUAAAAAGCAUUUGUUAA ...........((((.((...........((....))..((((......(((((((.((....)).))))))).....)))).)).)))).(((..(((((........)))))..))). ( -27.90) >consensus UAUUAUUUUCA____CUUAAUUUUAUUUCGCAAAUGCAGCUCGAAAAACGUUUGCUGUCCAGCGAAAGCAAACAAAAGCGAGCAACUGAGGUAACGAGUGCAAUAAAAACCAUUUGUUAA ...............((((..........((....)).(((((......(((((((.((....)).))))))).....)))))...)))).(((((((((..........))))))))). (-20.60 = -20.92 + 0.32)

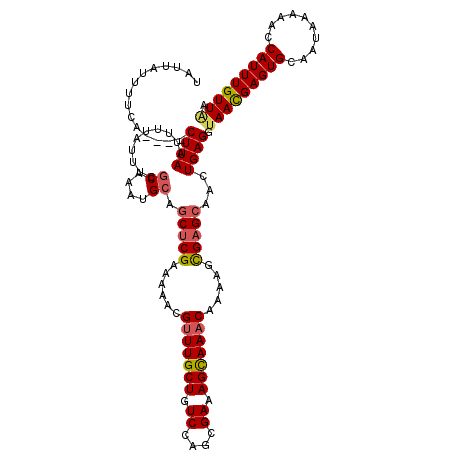
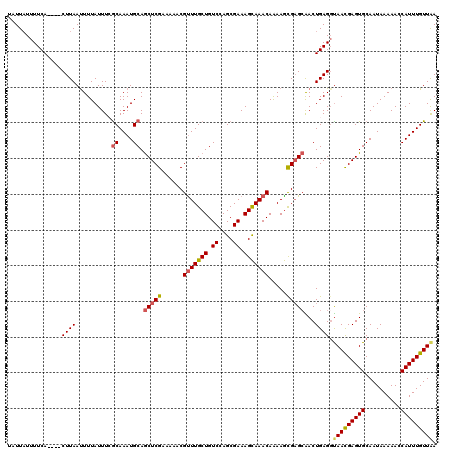
| Location | 17,652,557 – 17,652,675 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.73 |
| Mean single sequence MFE | -29.88 |
| Consensus MFE | -26.30 |
| Energy contribution | -26.22 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708315 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17652557 118 + 20766785 UCGAAAAACGUUUGCUGUCCAGCGAAAGCAAACAAAAGCGCGCAACUGAGGUAACGAGUGCAAUAAAAACCAUUUGUUAAUGACUUUUGAAUACGCAAUUUGCUGUUGUUGCUUGGGA-- .........(((((((.((....)).)))))))..(((((.(((((.(((.(((((((((..........)))))))))....)))........((.....)).))))))))))....-- ( -26.80) >DroSec_CAF1 6169 120 + 1 UCGAAAAACGUUUGCUGUCCAGCGAAAGUAAACAAAAACGAGCAACUGAGCUAACGAGUGCAAUAAAAACCAUUUGUUGAUGACUUUUGAAUACGCAAUUUGCUGUUGUUGCUUGGGUCC .........(((((((.((....)).))))))).....((((((((..((((((((((((..........))))))))).......(((......)))...)))...))))))))..... ( -27.30) >DroSim_CAF1 12067 120 + 1 UCGAAAAACGUUUGCUGUCCAGCGAAAGCAAACAAAAGCGAGCAACUGAGGUAACGAGUGCAAUAAAAACCAUUUGUUAAUGACUUUUGAAUACGCAAUUUGCUGUUGUUGCUUGGGUUC .........(((((((.....((....)).......)))))))((((.(((((((((..((((.......((((....))))....(((......))).))))..))))))))).)))). ( -31.00) >DroEre_CAF1 6302 119 + 1 UCGAAAAACGCUUGCUGUCCAGCGAAAGCAAACAGAAGUGAGCGACUGAGGCAAUGAGUGCAAUAAAAAGCAUUUGUUAAUGACUUUUGAAUACGCAAUUUGCUGUUGUUGCUUGGGUU- ........(((((.((((...((....))..)))))))))((((((...(((((...(((.....(((((((((....)))).))))).....)))...)))))...))))))......- ( -35.00) >DroYak_CAF1 17629 119 + 1 UCGAAAAACGUUUGCUGUCCAGCGAAAGCAAACAAAAGCGAGAGACUGAGGUAAUGAGUGCAAUAAAAAGCAUUUGUUAAUGACUUUUGAAUACGCAAUUUGCUGUUGUUGCUUGGGUU- (((......(((((((.((....)).))))))).....)))..((((.((((((..(..((((..(((((((((....)))).)))))...........))))..)..)))))).))))- ( -29.32) >consensus UCGAAAAACGUUUGCUGUCCAGCGAAAGCAAACAAAAGCGAGCAACUGAGGUAACGAGUGCAAUAAAAACCAUUUGUUAAUGACUUUUGAAUACGCAAUUUGCUGUUGUUGCUUGGGUU_ .........(((((((.((....)).))))))).....((((((((...(((((...(((...(((((..((((....))))..)))))....)))...)))))...))))))))..... (-26.30 = -26.22 + -0.08)

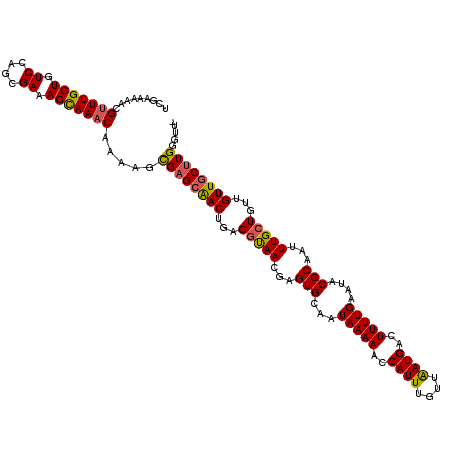
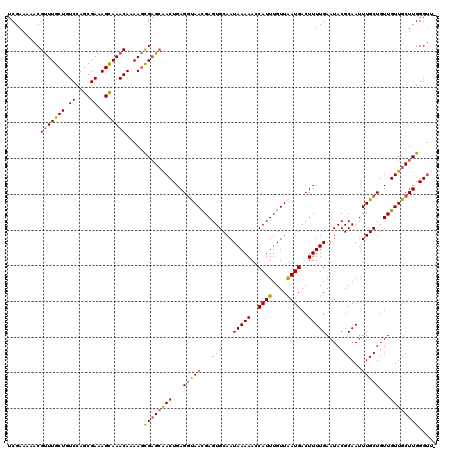
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:28 2006