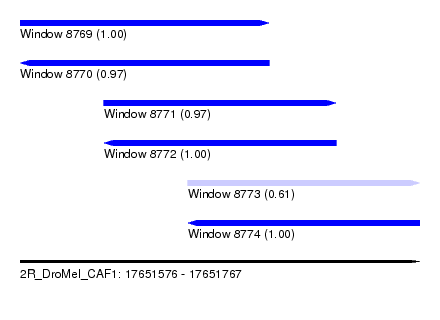
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,651,576 – 17,651,767 |
| Length | 191 |
| Max. P | 0.999153 |

| Location | 17,651,576 – 17,651,695 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.73 |
| Mean single sequence MFE | -29.33 |
| Consensus MFE | -27.57 |
| Energy contribution | -27.77 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.34 |
| SVM RNA-class probability | 0.999032 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17651576 119 + 20766785 UCAUAUUCGUAAUUAUUGUAAUGAGCCAUUUCUCUAGUUACUUUCCUCGUUGGCUGCAUGGGCUGGGUUUUUAAUUAAUUUUCCAUUGACCCAGCCUGACAGCUGAGCCUCAGCUUUU-U ........(((((((..(.((((...)))).)..)))))))..........(((((...((((((((((..................))))))))))..)))))((((....))))..-. ( -31.47) >DroSec_CAF1 5189 118 + 1 UCAUAUUCGUAAUUAUUGUAAUGAGCCAUUUCUCUAGUUACUUUCCUCGUUGGCCGC-UGGGCUGGGUUUUUAAUUAAUUUUCCAUUGACCCAACCUGACAGCUGAGCCUCAGCUUUU-U ........(((((((..(.((((...)))).)..))))))).......(((((((((-((((.((((((..................)))))).))...)))).).)))..)))....-. ( -24.17) >DroSim_CAF1 11092 119 + 1 UCAUAUUCGUAAUUAUUGUAAUGAGCCAUUUCUCUAGUUACUUUCCUCGUUGGCCGCAUGGGCUGGGUUUUUAAUUAAUUUUCCAUUGACCCAGCCUGACAGCUGAGCCUCAGCUUUU-C ........(((((((..(.((((...)))).)..)))))))...((.....))......((((((((((..................))))))))))...(((((.....)))))...-. ( -29.07) >DroEre_CAF1 5306 119 + 1 UCAUAUUCGUAAUUAUUGUAAUGAGCCAUUUCUCUAGUUACUUUCCUCGUUGGCCGUAUGGGCUGGGUUUUUAAUUAAUUUUCCAUUGACCCAGCCUGACAGCUGAGCCUCAGCUUUU-G .(((((..(((((((..(.((((...)))).)..)))))))...((.....))..)))))(((((((((..................))))))))).((.(((((.....))))).))-. ( -30.47) >DroYak_CAF1 16643 120 + 1 UCAUAUUCGUAAUUAUUGUAAUGAGCCAUUUCUCUAGUUACUUUCCUCGUUGAGCGGAUGGGCUGGGUUUUUAAUUAAUUUUCCAUUGACCCAGCCUGACAGCUGAGCCUCAGCUUUUUC ........(((((((..(.((((...)))).)..)))))))..(((.(.....).))).((((((((((..................))))))))))...(((((.....)))))..... ( -31.47) >consensus UCAUAUUCGUAAUUAUUGUAAUGAGCCAUUUCUCUAGUUACUUUCCUCGUUGGCCGCAUGGGCUGGGUUUUUAAUUAAUUUUCCAUUGACCCAGCCUGACAGCUGAGCCUCAGCUUUU_C ........(((((((..(.((((...)))).)..)))))))..................((((((((((..................))))))))))...(((((.....)))))..... (-27.57 = -27.77 + 0.20)

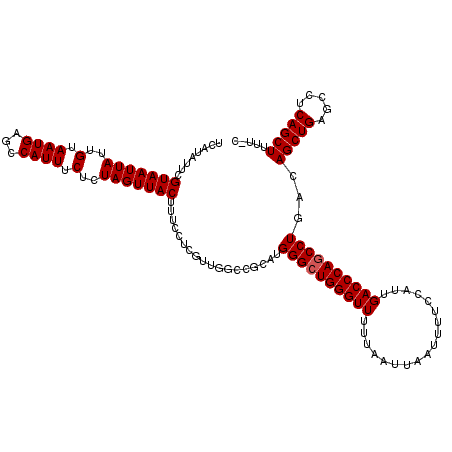
| Location | 17,651,576 – 17,651,695 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.73 |
| Mean single sequence MFE | -27.01 |
| Consensus MFE | -25.03 |
| Energy contribution | -24.83 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17651576 119 - 20766785 A-AAAAGCUGAGGCUCAGCUGUCAGGCUGGGUCAAUGGAAAAUUAAUUAAAAACCCAGCCCAUGCAGCCAACGAGGAAAGUAACUAGAGAAAUGGCUCAUUACAAUAAUUACGAAUAUGA .-...(((((...(((.(((((..((((((((....................))))))))...))))).....((........)).)))...)))))....................... ( -27.65) >DroSec_CAF1 5189 118 - 1 A-AAAAGCUGAGGCUCAGCUGUCAGGUUGGGUCAAUGGAAAAUUAAUUAAAAACCCAGCCCA-GCGGCCAACGAGGAAAGUAACUAGAGAAAUGGCUCAUUACAAUAAUUACGAAUAUGA .-...(((((...(((.(((((..((((((((....................))))))))..-))))).....((........)).)))...)))))....................... ( -26.05) >DroSim_CAF1 11092 119 - 1 G-AAAAGCUGAGGCUCAGCUGUCAGGCUGGGUCAAUGGAAAAUUAAUUAAAAACCCAGCCCAUGCGGCCAACGAGGAAAGUAACUAGAGAAAUGGCUCAUUACAAUAAUUACGAAUAUGA .-...(((((...(((.(((((..((((((((....................))))))))...))))).....((........)).)))...)))))....................... ( -26.95) >DroEre_CAF1 5306 119 - 1 C-AAAAGCUGAGGCUCAGCUGUCAGGCUGGGUCAAUGGAAAAUUAAUUAAAAACCCAGCCCAUACGGCCAACGAGGAAAGUAACUAGAGAAAUGGCUCAUUACAAUAAUUACGAAUAUGA .-...(((((.....)))))((((((((((((....................))))))))..(((..((.....))...)))..........))))........................ ( -26.85) >DroYak_CAF1 16643 120 - 1 GAAAAAGCUGAGGCUCAGCUGUCAGGCUGGGUCAAUGGAAAAUUAAUUAAAAACCCAGCCCAUCCGCUCAACGAGGAAAGUAACUAGAGAAAUGGCUCAUUACAAUAAUUACGAAUAUGA ((...(((((.....))))).)).((((((((....................))))))))..(((.(.....).)))..((((...(((......))).))))................. ( -27.55) >consensus A_AAAAGCUGAGGCUCAGCUGUCAGGCUGGGUCAAUGGAAAAUUAAUUAAAAACCCAGCCCAUGCGGCCAACGAGGAAAGUAACUAGAGAAAUGGCUCAUUACAAUAAUUACGAAUAUGA .....(((((...(((.(((((..((((((((....................))))))))...))))).....((........)).)))...)))))....................... (-25.03 = -24.83 + -0.20)

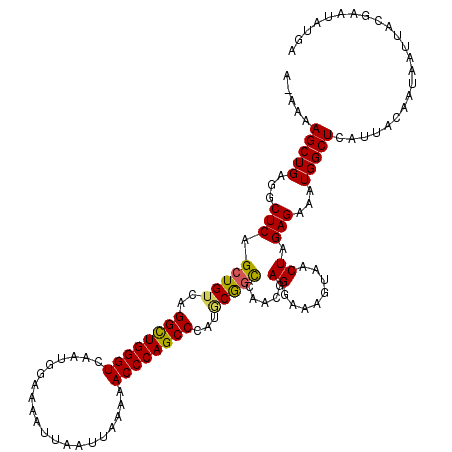
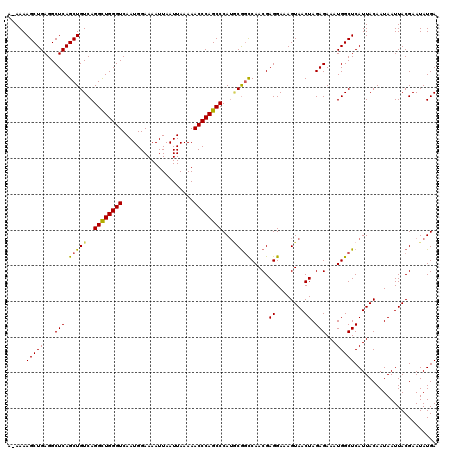
| Location | 17,651,616 – 17,651,727 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.41 |
| Mean single sequence MFE | -36.96 |
| Consensus MFE | -25.97 |
| Energy contribution | -27.13 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.70 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17651616 111 + 20766785 CUUUCCUCGUUGGCUGCAUGGGCUGGGUUUUUAAUUAAUUUUCCAUUGACCCAGCCUGACAGCUGAGCCUCAGCUUUU-UCCUCUUUUAUU--UCUGAGCUGAGCUCAGAG------GCU ....((((...(((((...((((((((((..................))))))))))..)))))((((.(((((((..-............--...))))))))))).)))------).. ( -44.50) >DroSec_CAF1 5229 116 + 1 CUUUCCUCGUUGGCCGC-UGGGCUGGGUUUUUAAUUAAUUUUCCAUUGACCCAACCUGACAGCUGAGCCUCAGCUUUU-UCCUCUUUUAUU--UCUGAGCCGAGCUCAGAGCUCAGAGCU ...........(((..(-(((((((((((..................)))))).......(((((.....)))))...-............--(((((((...))))))))))))).))) ( -32.87) >DroSim_CAF1 11132 110 + 1 CUUUCCUCGUUGGCCGCAUGGGCUGGGUUUUUAAUUAAUUUUCCAUUGACCCAGCCUGACAGCUGAGCCUCAGCUUUU-CCCUCUUUUAUU--UCUGAGCCGAGCUC-------GGAGCU ....((.....))..((..((((((((((..................))))))))))...(((((.....)))))...-............--(((((((...))))-------))))). ( -33.87) >DroEre_CAF1 5346 110 + 1 CUUUCCUCGUUGGCCGUAUGGGCUGGGUUUUUAAUUAAUUUUCCAUUGACCCAGCCUGACAGCUGAGCCUCAGCUUUU-GCCUCUUUUAUU--UUUGAGCUGAGCUC-------AGAGCC ...........(((.((.(((((((((((..................)))))))))))))..((((((.(((((((..-............--...)))))))))))-------)).))) ( -40.10) >DroYak_CAF1 16683 113 + 1 CUUUCCUCGUUGAGCGGAUGGGCUGGGUUUUUAAUUAAUUUUCCAUUGACCCAGCCUGACAGCUGAGCCUCAGCUUUUUCCCUCUUUUAUUUGUUUGAGCCCAGAUC-------AGAGCU .....(((...(((.(((.((((((((((..................))))))))))...(((((.....)))))...))))))........(((((....))))).-------.))).. ( -33.47) >consensus CUUUCCUCGUUGGCCGCAUGGGCUGGGUUUUUAAUUAAUUUUCCAUUGACCCAGCCUGACAGCUGAGCCUCAGCUUUU_CCCUCUUUUAUU__UCUGAGCCGAGCUC_______AGAGCU .........(((((.....((((((((((..................))))))))))...(((((.....))))).......................)))))((((........)))). (-25.97 = -27.13 + 1.16)

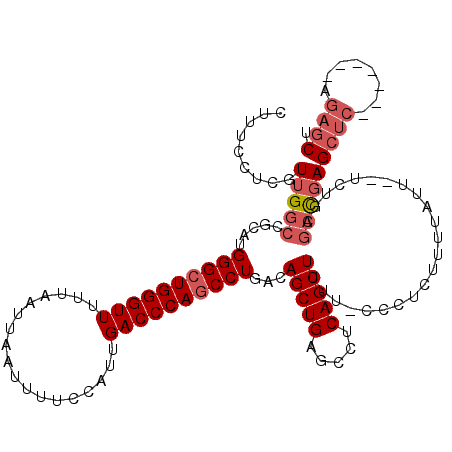
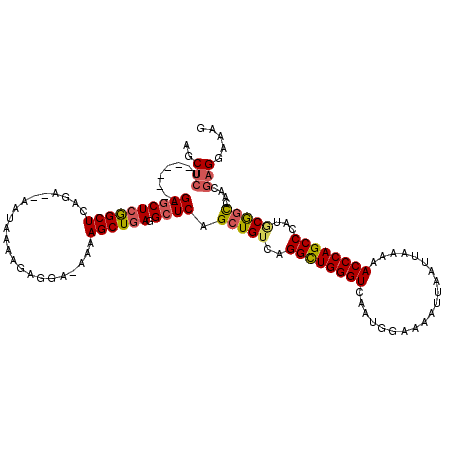
| Location | 17,651,616 – 17,651,727 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.41 |
| Mean single sequence MFE | -36.78 |
| Consensus MFE | -29.66 |
| Energy contribution | -30.02 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.72 |
| Structure conservation index | 0.81 |
| SVM decision value | 3.40 |
| SVM RNA-class probability | 0.999153 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17651616 111 - 20766785 AGC------CUCUGAGCUCAGCUCAGA--AAUAAAAGAGGA-AAAAGCUGAGGCUCAGCUGUCAGGCUGGGUCAAUGGAAAAUUAAUUAAAAACCCAGCCCAUGCAGCCAACGAGGAAAG ..(------((((((((((((((....--............-...)))))).)))))(((((..((((((((....................))))))))...)))))....)))).... ( -40.56) >DroSec_CAF1 5229 116 - 1 AGCUCUGAGCUCUGAGCUCGGCUCAGA--AAUAAAAGAGGA-AAAAGCUGAGGCUCAGCUGUCAGGUUGGGUCAAUGGAAAAUUAAUUAAAAACCCAGCCCA-GCGGCCAACGAGGAAAG ..((((((((.((.((((...(((...--.......)))..-...)))).)))))))(((((..((((((((....................))))))))..-)))))....)))..... ( -37.55) >DroSim_CAF1 11132 110 - 1 AGCUCC-------GAGCUCGGCUCAGA--AAUAAAAGAGGG-AAAAGCUGAGGCUCAGCUGUCAGGCUGGGUCAAUGGAAAAUUAAUUAAAAACCCAGCCCAUGCGGCCAACGAGGAAAG ...(((-------((((((((((....--............-...)))))).)))).(((((..((((((((....................))))))))...)))))......)))... ( -36.06) >DroEre_CAF1 5346 110 - 1 GGCUCU-------GAGCUCAGCUCAAA--AAUAAAAGAGGC-AAAAGCUGAGGCUCAGCUGUCAGGCUGGGUCAAUGGAAAAUUAAUUAAAAACCCAGCCCAUACGGCCAACGAGGAAAG ..((((-------((((((((((....--............-...)))))).)))))(((((..((((((((....................))))))))...)))))....)))..... ( -36.46) >DroYak_CAF1 16683 113 - 1 AGCUCU-------GAUCUGGGCUCAAACAAAUAAAAGAGGGAAAAAGCUGAGGCUCAGCUGUCAGGCUGGGUCAAUGGAAAAUUAAUUAAAAACCCAGCCCAUCCGCUCAACGAGGAAAG (((((.-------.....))))).............((((((...(((((.....)))))....((((((((....................))))))))..))).)))........... ( -33.25) >consensus AGCUCU_______GAGCUCGGCUCAGA__AAUAAAAGAGGA_AAAAGCUGAGGCUCAGCUGUCAGGCUGGGUCAAUGGAAAAUUAAUUAAAAACCCAGCCCAUGCGGCCAACGAGGAAAG ..(((........((((((((((......................)))))).)))).(((((..((((((((....................))))))))...)))))....)))..... (-29.66 = -30.02 + 0.36)

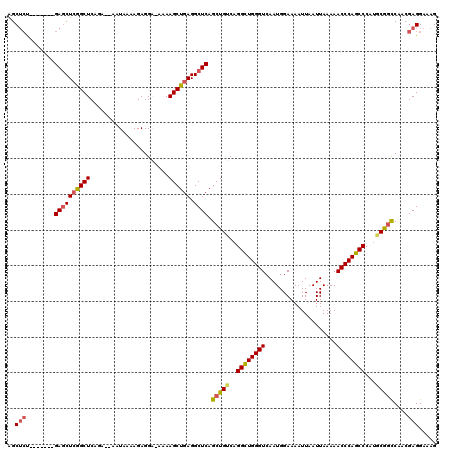
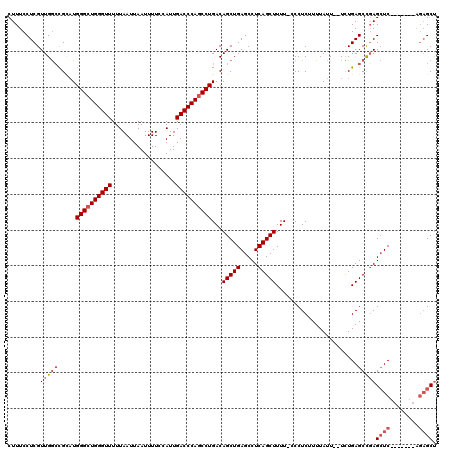
| Location | 17,651,656 – 17,651,767 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.41 |
| Mean single sequence MFE | -23.84 |
| Consensus MFE | -17.44 |
| Energy contribution | -18.24 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.613787 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17651656 111 + 20766785 UUCCAUUGACCCAGCCUGACAGCUGAGCCUCAGCUUUU-UCCUCUUUUAUU--UCUGAGCUGAGCUCAGAG------GCUCUGCCAGCCCAAGAGCAUUUCAUAUUAAUUCUCAUUUUUU .....(((.(..(((((.....((((((.(((((((..-............--...)))))))))))))))------)))..).))).....(((.(((.......))).)))....... ( -26.63) >DroSec_CAF1 5268 117 + 1 UUCCAUUGACCCAACCUGACAGCUGAGCCUCAGCUUUU-UCCUCUUUUAUU--UCUGAGCCGAGCUCAGAGCUCAGAGCUCUGCCAGCCAGAGAGCAUUUCAUAUUAAUUCUCAUUUUUU ......(((...........(((((.....)))))...-..((((((....--.(((.((.((((((........)))))).)))))..))))))....))).................. ( -26.30) >DroSim_CAF1 11172 110 + 1 UUCCAUUGACCCAGCCUGACAGCUGAGCCUCAGCUUUU-CCCUCUUUUAUU--UCUGAGCCGAGCUC-------GGAGCUCUGCCAGCCCAAGAGCAUUUCAUAUUAAUUCUCAUUUUUU .(((........(((......)))((((.((.((((..-............--...)))).))))))-------)))(((((.........)))))........................ ( -22.53) >DroEre_CAF1 5386 110 + 1 UUCCAUUGACCCAGCCUGACAGCUGAGCCUCAGCUUUU-GCCUCUUUUAUU--UUUGAGCUGAGCUC-------AGAGCCAUGCCAGCCCAAGAGCAUUUCAUAUUAAUUCCCAUUUUUU ......(((....((.((....((((((.(((((((..-............--...)))))))))))-------))...)).))..((......))...))).................. ( -22.63) >DroYak_CAF1 16723 113 + 1 UUCCAUUGACCCAGCCUGACAGCUGAGCCUCAGCUUUUUCCCUCUUUUAUUUGUUUGAGCCCAGAUC-------AGAGCUUGGCCAGCCCAAGAGCAUUUCAUAUUAAUUCUCAUUUUUU .............((((....((((.(((..((((((.((.(((............)))....))..-------)))))).)))))))...)).))........................ ( -21.10) >consensus UUCCAUUGACCCAGCCUGACAGCUGAGCCUCAGCUUUU_CCCUCUUUUAUU__UCUGAGCCGAGCUC_______AGAGCUCUGCCAGCCCAAGAGCAUUUCAUAUUAAUUCUCAUUUUUU ......(((...........(((((.....)))))......(((((........(((.((.((((((........)))))).)))))...)))))....))).................. (-17.44 = -18.24 + 0.80)

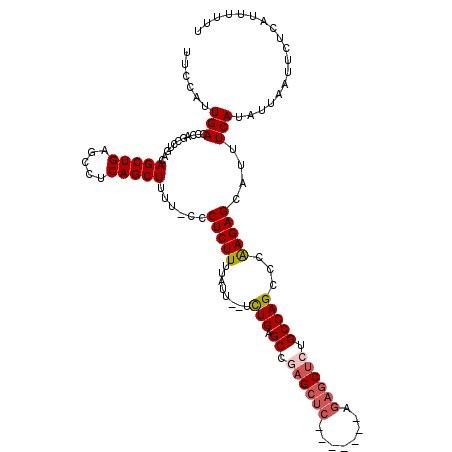
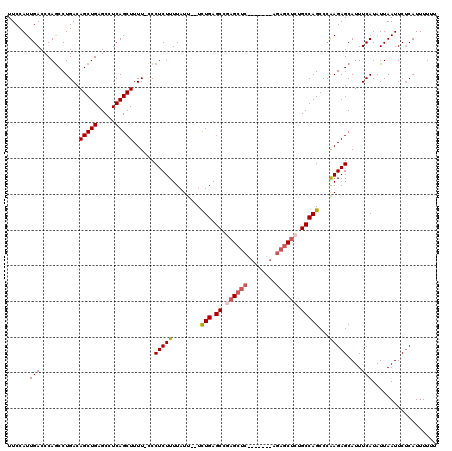
| Location | 17,651,656 – 17,651,767 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.41 |
| Mean single sequence MFE | -32.40 |
| Consensus MFE | -27.02 |
| Energy contribution | -28.30 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.95 |
| SVM RNA-class probability | 0.997875 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17651656 111 - 20766785 AAAAAAUGAGAAUUAAUAUGAAAUGCUCUUGGGCUGGCAGAGC------CUCUGAGCUCAGCUCAGA--AAUAAAAGAGGA-AAAAGCUGAGGCUCAGCUGUCAGGCUGGGUCAAUGGAA ........................((((..(((((.....)))------))..))))((((((....--............-...))))))(((((((((....)))))))))....... ( -34.11) >DroSec_CAF1 5268 117 - 1 AAAAAAUGAGAAUUAAUAUGAAAUGCUCUCUGGCUGGCAGAGCUCUGAGCUCUGAGCUCGGCUCAGA--AAUAAAAGAGGA-AAAAGCUGAGGCUCAGCUGUCAGGUUGGGUCAAUGGAA ........................(((((((..(((((.((((((........)))))).)).))).--......))))..-...)))...(((((((((....)))))))))....... ( -32.90) >DroSim_CAF1 11172 110 - 1 AAAAAAUGAGAAUUAAUAUGAAAUGCUCUUGGGCUGGCAGAGCUCC-------GAGCUCGGCUCAGA--AAUAAAAGAGGG-AAAAGCUGAGGCUCAGCUGUCAGGCUGGGUCAAUGGAA .......................(.(((((...(((((.((((...-------..)))).)).))).--.....))))).)-.........(((((((((....)))))))))....... ( -31.80) >DroEre_CAF1 5386 110 - 1 AAAAAAUGGGAAUUAAUAUGAAAUGCUCUUGGGCUGGCAUGGCUCU-------GAGCUCAGCUCAAA--AAUAAAAGAGGC-AAAAGCUGAGGCUCAGCUGUCAGGCUGGGUCAAUGGAA .........................((.((((.(..((.((((.((-------((((((((((....--............-...)))))).))))))..)))).))..).)))).)).. ( -33.81) >DroYak_CAF1 16723 113 - 1 AAAAAAUGAGAAUUAAUAUGAAAUGCUCUUGGGCUGGCCAAGCUCU-------GAUCUGGGCUCAAACAAAUAAAAGAGGGAAAAAGCUGAGGCUCAGCUGUCAGGCUGGGUCAAUGGAA .........................((.((((.(..(((.(((((.-------.....))))).................((...(((((.....))))).)).)))..).)))).)).. ( -29.40) >consensus AAAAAAUGAGAAUUAAUAUGAAAUGCUCUUGGGCUGGCAGAGCUCU_______GAGCUCGGCUCAGA__AAUAAAAGAGGA_AAAAGCUGAGGCUCAGCUGUCAGGCUGGGUCAAUGGAA .........................(((((...(((((.((((((........)))))).)).)))........)))))............(((((((((....)))))))))....... (-27.02 = -28.30 + 1.28)

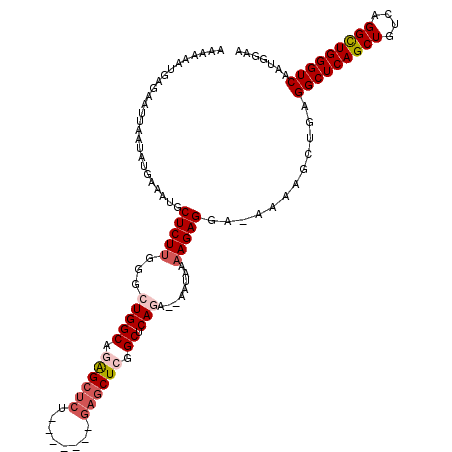
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:25 2006