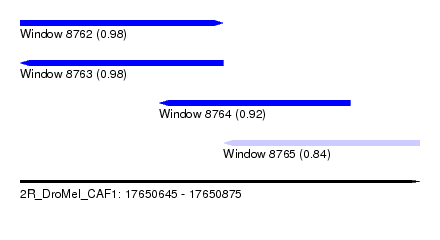
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,650,645 – 17,650,875 |
| Length | 230 |
| Max. P | 0.984607 |

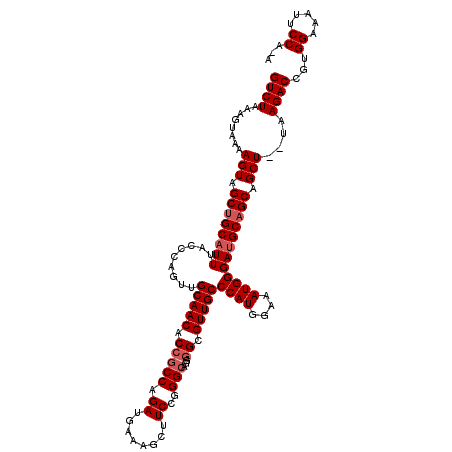
| Location | 17,650,645 – 17,650,762 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.09 |
| Mean single sequence MFE | -38.98 |
| Consensus MFE | -32.36 |
| Energy contribution | -32.80 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17650645 117 + 20766785 GUCUAAAGUAAAAGUAGCUGCAUUUACCUAGUUGCAACACCGCCAGAUGAAAGCUUCCGGGCAGGGGCGUUGCCCAUGGAAAUGGGAUGCAGCAGCU--UAAGACGGUGGAAAUUCCA-A .((((..((..((((.((((((((.........(((((.(((((.((........))..)))...)).)))))((((....)))))))))))).)))--)...))..)))).......-. ( -39.40) >DroSec_CAF1 4243 117 + 1 GUCUAAAGUAAAAGUAGCUGCAUUUACCCAGUUGCAACACCGCCAGAUGAAAGCUUCAGGGAAGGGGCGUUGCCCAUGGAAAUGGGAUGCAGCAGCU--UAAGACCGUGGAAAUUCCA-A .((((..((..((((.((((((((.........(((((.((.((...(((.....))).....)))).)))))((((....)))))))))))).)))--)...))..)))).......-. ( -37.80) >DroSim_CAF1 10161 117 + 1 GUCUAAAGUAAAAGUAGCUGCAUUUACCCAGUUGCAACACCGCCAGAUGAAAGCUUCCGGGCAGGGGCGUUGCCCAUGGAAAUGGGAUGCAGCAGCU--UAAGACCGUGGAAAUUCCA-A .((((..((..((((.((((((((.........(((((.(((((.((........))..)))...)).)))))((((....)))))))))))).)))--)...))..)))).......-. ( -38.50) >DroEre_CAF1 4306 117 + 1 GUCUAAAGUAAAAGUAGCUGCAUUUACCCAGUUGCAACACAGCCAGAUGAAAGCUUCCGGGCAGGGGCGUUGCCCAUGGAAAUGGGAUGCAGCAGCU--UAAGACCAAGGAAAUUCCA-A ((((.((((.......(((((((...((((((((.....))))...........((((((((((.....))))))..)))).))))))))))).)))--).))))...((.....)).-. ( -35.50) >DroYak_CAF1 15580 120 + 1 GUCUAAAGUAAAAGUAGCUGCAUUUACCCAGUUGCAACACCGCCAGAUGAAAGCUUCCGGGCAGGGGCGUUGCCCAUGGAAAUGGGACGCUGCUGCUGCUUAGACCGAGGAAAUUCCAAA ((((((.(((...(((((((........)))))))....(((..((.(....)))..)))((((.(((((..(((((....)))))))))).)))))))))))))...((.....))... ( -43.70) >consensus GUCUAAAGUAAAAGUAGCUGCAUUUACCCAGUUGCAACACCGCCAGAUGAAAGCUUCCGGGCAGGGGCGUUGCCCAUGGAAAUGGGAUGCAGCAGCU__UAAGACCGUGGAAAUUCCA_A ((((........(((.(((((((...((((.((.((.....(((.((........))..)))..((((...)))).)).)).))))))))))).)))....))))...((.....))... (-32.36 = -32.80 + 0.44)

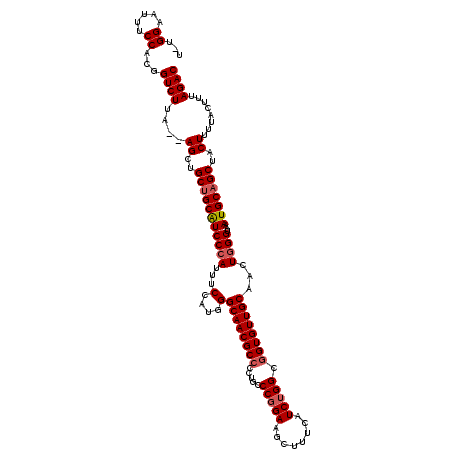
| Location | 17,650,645 – 17,650,762 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.09 |
| Mean single sequence MFE | -36.38 |
| Consensus MFE | -31.26 |
| Energy contribution | -31.90 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17650645 117 - 20766785 U-UGGAAUUUCCACCGUCUUA--AGCUGCUGCAUCCCAUUUCCAUGGGCAACGCCCCUGCCCGGAAGCUUUCAUCUGGCGGUGUUGCAACUAGGUAAAUGCAGCUACUUUUACUUUAGAC .-(((.....)))..((((.(--((..(((((((.((((....))))((((((((...(((.(((........))))))))))))))..........)))))))..))).......)))) ( -36.40) >DroSec_CAF1 4243 117 - 1 U-UGGAAUUUCCACGGUCUUA--AGCUGCUGCAUCCCAUUUCCAUGGGCAACGCCCCUUCCCUGAAGCUUUCAUCUGGCGGUGUUGCAACUGGGUAAAUGCAGCUACUUUUACUUUAGAC .-(((.....)))..((((.(--((..(((((((((((...(....)((((((((((.....(((.....)))...)).))))))))...))))...)))))))..))).......)))) ( -35.90) >DroSim_CAF1 10161 117 - 1 U-UGGAAUUUCCACGGUCUUA--AGCUGCUGCAUCCCAUUUCCAUGGGCAACGCCCCUGCCCGGAAGCUUUCAUCUGGCGGUGUUGCAACUGGGUAAAUGCAGCUACUUUUACUUUAGAC .-(((.....)))..((((.(--((..(((((((...........((((...)))).(((((((..((...((((....))))..))..))))))).)))))))..))).......)))) ( -37.60) >DroEre_CAF1 4306 117 - 1 U-UGGAAUUUCCUUGGUCUUA--AGCUGCUGCAUCCCAUUUCCAUGGGCAACGCCCCUGCCCGGAAGCUUUCAUCUGGCUGUGUUGCAACUGGGUAAAUGCAGCUACUUUUACUUUAGAC .-.((.....))...((((.(--((..(((((((...........((((...)))).(((((((..((...(((......)))..))..))))))).)))))))..))).......)))) ( -33.80) >DroYak_CAF1 15580 120 - 1 UUUGGAAUUUCCUCGGUCUAAGCAGCAGCAGCGUCCCAUUUCCAUGGGCAACGCCCCUGCCCGGAAGCUUUCAUCUGGCGGUGUUGCAACUGGGUAAAUGCAGCUACUUUUACUUUAGAC ...((.....))..(((..(((.(((.((((((((((((....)))))..))))...(((((((..((...((((....))))..))..)))))))..))).))).)))..)))...... ( -38.20) >consensus U_UGGAAUUUCCACGGUCUUA__AGCUGCUGCAUCCCAUUUCCAUGGGCAACGCCCCUGCCCGGAAGCUUUCAUCUGGCGGUGUUGCAACUGGGUAAAUGCAGCUACUUUUACUUUAGAC ...((.....))...((((....((..(((((((((((...(....)((((((((.....(((((........))))).))))))))...))))...)))))))..))........)))) (-31.26 = -31.90 + 0.64)

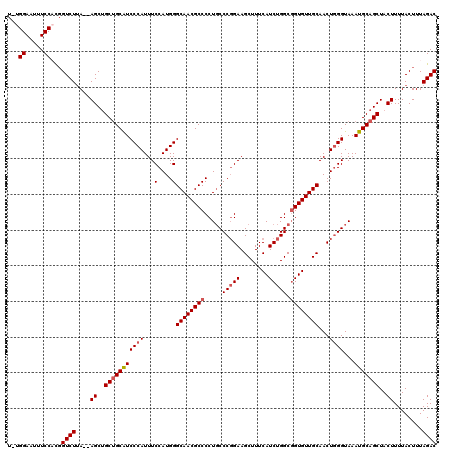
| Location | 17,650,725 – 17,650,835 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.56 |
| Mean single sequence MFE | -34.18 |
| Consensus MFE | -25.10 |
| Energy contribution | -24.82 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.917222 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17650725 110 - 20766785 AGAUAUAUGCUUUGUUUUAAGCGCA-------GCAGAUGGAAAUCCGAGGGGAUUUUCAUGAAAAUGUGUUCCAGUUACUU-UGGAAUUUCCACCGUCUUA--AGCUGCUGCAUCCCAUU .(((....((((......))))(((-------((((((((((((((....))))))))))((...((.(((((((.....)-))))))...))...))...--..))))))))))..... ( -35.90) >DroSec_CAF1 4323 110 - 1 AGAUAAAUGCUUUGUCUUAAGCGGA-------GGAGAUGGAAAUCCGAGGGGAUUUUCAUGAAAAUGUGUUCCAGUUACUU-UGGAAUUUCCACGGUCUUA--AGCUGCUGCAUCCCAUU ......((((...(.(((((((.(.-------((((((((((((((....))))))))))........(((((((.....)-)))))))))).).).))))--)))....))))...... ( -32.10) >DroSim_CAF1 10241 110 - 1 AGAUAAAUGCUUUGUCUUAAGCGAA-------GCAGAUGGAAAUCCGAGGGGAUUUUCAUGAAAAUGUGUUCCAGUUACUU-UGGAAUUUCCACGGUCUUA--AGCUGCUGCAUCCCAUU .......((((((((.....)))))-------))).((((((((((....))))))))))......((((..((((((((.-(((.....))).)))....--)))))..))))...... ( -32.10) >DroEre_CAF1 4386 110 - 1 AGAUAAACGCUUUGUUUUAGGCGAG-------GCAGAUGGAAAUCCGAGGGGAUUUUCAUGAAAAUGAGUUCCAGUUACUU-UGGAAUUUCCUUGGUCUUA--AGCUGCUGCAUCCCAUU ........(((((((.....)))))-------)).(((((....((((((..(..(((((....)))))((((((.....)-))))))..)))))).....--.((....))...))))) ( -34.70) >DroYak_CAF1 15660 120 - 1 AGAUAAAUGCUUUGUUUUAAGCCAGACAGAUGGCAGAUGGAAAUCCAAGGGGAUUUUCAUGAAAAUGAGUUCCAGUUACUUUUGGAAUUUCCUCGGUCUAAGCAGCAGCAGCGUCCCAUU .(((...((((((((((...((((......))))((((((....))..(((((...((((....))))(((((((......))))))).))))).)))))))))).))))..)))..... ( -36.10) >consensus AGAUAAAUGCUUUGUUUUAAGCGAA_______GCAGAUGGAAAUCCGAGGGGAUUUUCAUGAAAAUGUGUUCCAGUUACUU_UGGAAUUUCCACGGUCUUA__AGCUGCUGCAUCCCAUU .(((...(((((......))))).........((((((((((((((....))))))))))((...(((((((((........)))))....)))).))..........)))))))..... (-25.10 = -24.82 + -0.28)

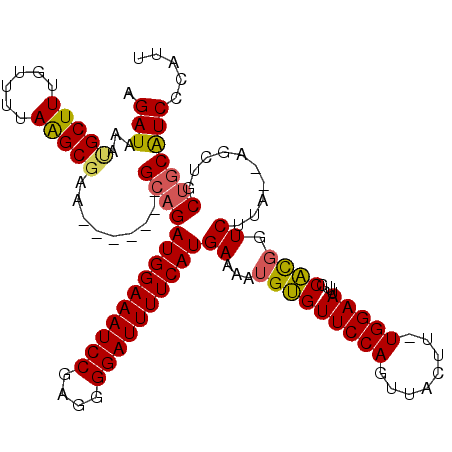
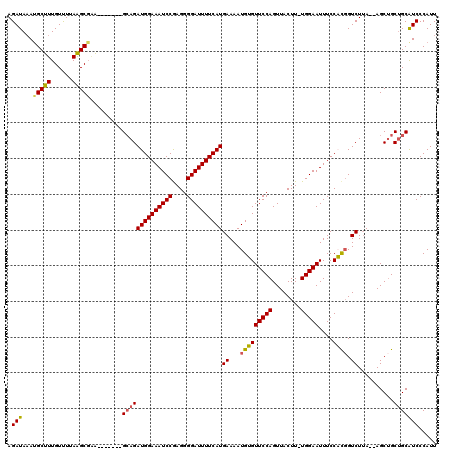
| Location | 17,650,762 – 17,650,875 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.37 |
| Mean single sequence MFE | -29.36 |
| Consensus MFE | -22.74 |
| Energy contribution | -22.82 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.836535 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17650762 113 - 20766785 AAAUUCGGUUGUAAACGCAUAUUUGCUGGAGAUAAAUGUAAGAUAUAUGCUUUGUUUUAAGCGCA-------GCAGAUGGAAAUCCGAGGGGAUUUUCAUGAAAAUGUGUUCCAGUUACU ......((.....(((((((((((((((.......(((((...)))))((((......)))).))-------))))))((((((((....))))))))......)))))))))....... ( -28.60) >DroSec_CAF1 4360 112 - 1 -AACUCAAUUGUAAACGCAUAUUUGCUGGCGAUAAAUGUAAGAUAAAUGCUUUGUCUUAAGCGGA-------GGAGAUGGAAAUCCGAGGGGAUUUUCAUGAAAAUGUGUUCCAGUUACU -.....(((((..(((((((.((((((...((((((.(((.......)))))))))...))))))-------....((((((((((....))))))))))....))))))).)))))... ( -30.00) >DroSim_CAF1 10278 112 - 1 -AACUCGGUUGUAAACGCAUAUUUGCUGGCGAUAAAUGUAAGAUAAAUGCUUUGUCUUAAGCGAA-------GCAGAUGGAAAUCCGAGGGGAUUUUCAUGAAAAUGUGUUCCAGUUACU -((((.((.....((((((((((((((.((........((((((((.....)))))))).))..)-------))))))((((((((....))))))))......)))))))))))))... ( -31.30) >DroEre_CAF1 4423 113 - 1 AAACUCCGUUGAAAACGCGUAUUUGCUAGCGAUAAAUGUAAGAUAAACGCUUUGUUUUAGGCGAG-------GCAGAUGGAAAUCCGAGGGGAUUUUCAUGAAAAUGAGUUCCAGUUACU .(((((.(((......((((..(((....)))...))))......)))(((((((.....)))))-------))..((((((((((....))))))))))......)))))......... ( -26.70) >DroYak_CAF1 15700 120 - 1 GAACUCCGUUGAAAACGCAUAUUUGCUAGAGAUAAAUGUAAGAUAAAUGCUUUGUUUUAAGCCAGACAGAUGGCAGAUGGAAAUCCAAGGGGAUUUUCAUGAAAAUGAGUUCCAGUUACU ((((((..........(((....)))..((((((((.(((.......)))))))))))..((((......))))..((((((((((....))))))))))......))))))........ ( -30.20) >consensus _AACUCCGUUGUAAACGCAUAUUUGCUGGCGAUAAAUGUAAGAUAAAUGCUUUGUUUUAAGCGAA_______GCAGAUGGAAAUCCGAGGGGAUUUUCAUGAAAAUGUGUUCCAGUUACU .............(((((((.((((((...((((((.(((.......)))))))))...))))))...........((((((((((....))))))))))....)))))))......... (-22.74 = -22.82 + 0.08)

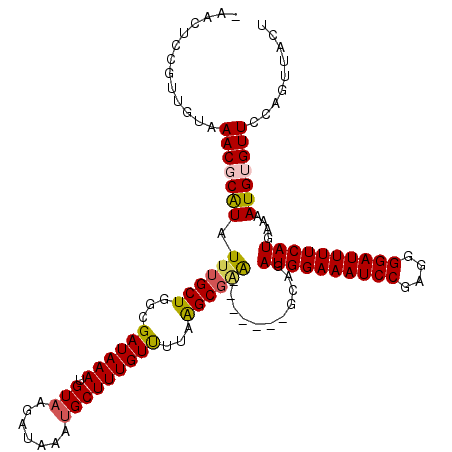
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:16 2006