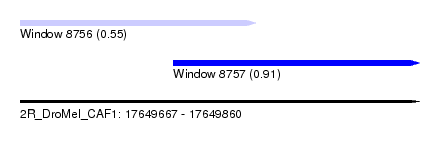
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,649,667 – 17,649,860 |
| Length | 193 |
| Max. P | 0.908918 |

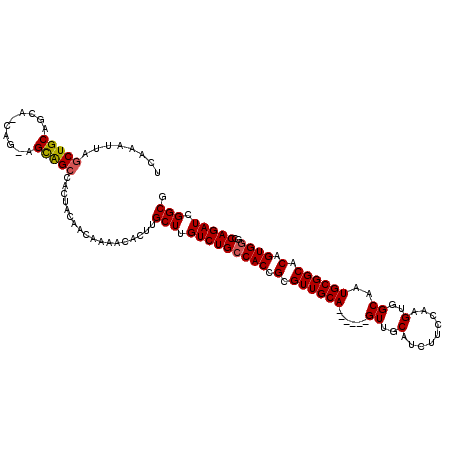
| Location | 17,649,667 – 17,649,781 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.40 |
| Mean single sequence MFE | -37.26 |
| Consensus MFE | -29.94 |
| Energy contribution | -30.02 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.553870 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17649667 114 + 20766785 UCAAAUUAGCUGCAGCAGCAGCAGUGGGCACUACAACAAAACACUUGCUUGUCUGCCACCGCGUUGCA------GUUGCAUCUUCCAAGUGGCAAUGCGGCACAGUGGGCCAGAUCGGCG ........(((((....))))).(((((((..(((((((.....))).)))).)))).((((((((((------.(((.......))).).)))))))))))).....(((.....))). ( -40.20) >DroSec_CAF1 3284 114 + 1 UCAAAUUAGCUGCAGCACCAGGAGCAGCCACUACAACAAAACACUUGCUUGUCUGCCACCGCGUUGCA------GUUGCAUCUUCCAAGUGGCAAUGCGGCACAGUGGGCCAGAUCGGCG ........(((((..........)))))..................(((.(((((((((.(.((((((------.((((..((....))..)))))))))).).))))..))))).))). ( -34.80) >DroSim_CAF1 9202 114 + 1 UCAAAUUAGCUGCAGCACCAGCAGCAGCCACUACAACAAAACACUUGCUUGUCUGCCACCGCGUUGCA------GUUGCAUCUUCCAAGUGGCAAUGCGGCACAGUGGGCCAGAUCGGCG ........(((((.((....)).)))))..................(((.(((((((((.(.((((((------.((((..((....))..)))))))))).).))))..))))).))). ( -38.00) >DroEre_CAF1 3375 108 + 1 UCAAAUUAGCUGCAGCA------GCAGCCACUACAACAAAACACUUGCUUGUCUGCCACCGCGUUGCA------GUUGCAUCUUCCAAGUGGCAAUGCGGCACAGUGGGCCAGAUCGGCG ........(((((....------)))))..................(((.(((((((((.(.((((((------.((((..((....))..)))))))))).).))))..))))).))). ( -35.70) >DroYak_CAF1 14615 114 + 1 UCAAAUUAGCUGCAGCA------GCAGCGACUACAACAAAACACUUGCUUGUCUGACACCGCGUUGCAGUUGCAGUUGCAUCUUCCAAGUGGCAAUGCGGCACAGUGGGCCAGAUCGGCA ........(((((....------))))).................((((.(((((.(.((((..(((....)))(((((((...((....))..)))))))...))))).))))).)))) ( -37.60) >consensus UCAAAUUAGCUGCAGCA_CAG_AGCAGCCACUACAACAAAACACUUGCUUGUCUGCCACCGCGUUGCA______GUUGCAUCUUCCAAGUGGCAAUGCGGCACAGUGGGCCAGAUCGGCG ........(((((..........)))))..................(((.(((((((((.(.((((((......((..(.........)..))..)))))).).))))..))))).))). (-29.94 = -30.02 + 0.08)


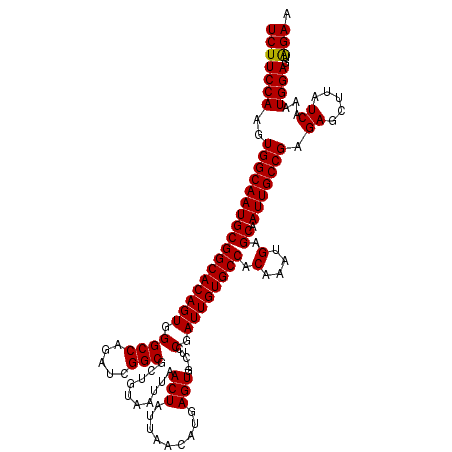
| Location | 17,649,741 – 17,649,860 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.57 |
| Mean single sequence MFE | -36.81 |
| Consensus MFE | -32.62 |
| Energy contribution | -32.46 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908918 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17649741 119 + 20766785 UCUUCCAAGUGGCAAUGCGGCACAGUGGGCCAGAUCGGCGCUGUAAUUAACUAUUAACACGAGUG-CUCCGAUUGUGCCACAAAUGAGCAAUUGCCGAGAGCUUAUCAAAUGGAGUAGAA (((((((..(((((((((((((((((.((.......((((((.((((.....)))).....))))-)))).)))))))).(....).)).))))))).((.....))...))))..))). ( -36.11) >DroSec_CAF1 3358 119 + 1 UCUUCCAAGUGGCAAUGCGGCACAGUGGGCCAGAUCGGCGCCGUAAUUAACUAUUAACAUGAGUG-CUCCGAUUGUGCCACAAAUGAGCAAUUGCCGAGAGCUUAUCAAAUGGAGUAGAA (((((((..(((((((((((((((((.((.......((((((((..............))).)))-)))).)))))))).(....).)).))))))).((.....))...))))..))). ( -35.55) >DroSim_CAF1 9276 119 + 1 UCUUCCAAGUGGCAAUGCGGCACAGUGGGCCAGAUCGGCGCUGUAAUUAACUAUUAACAUGAGUG-CUCCGAUUGUGCCACAAAUGAGCAAUUGCCGAGAGCUUAUCAAAUGGAGUAGAA (((((((..(((((((((((((((((.((.......((((((.((((.....)))).....))))-)))).)))))))).(....).)).))))))).((.....))...))))..))). ( -36.11) >DroEre_CAF1 3443 119 + 1 UCUUCCAAGUGGCAAUGCGGCACAGUGGGCCAGAUCGGCGCGGCAAUUAACUAUUACCAUGAGUG-CUGCGAUUGUGCCACAAAUGAGCAAUUGCCGAGAGCUUAUCAAAUGGAGUGGAA (((((((.((((((.((((((((.(((((((.....)))..(........).....))))..)))-)))))....))))))..((((((...........))))))....))))..))). ( -40.70) >DroYak_CAF1 14689 120 + 1 UCUUCCAAGUGGCAAUGCGGCACAGUGGGCCAGAUCGGCAGUGUAAUUAACUAUUAACAUGAGUGGCUGCGAUUGUGCCACAAAUGAGCAAUUGCCGAGAGCUUAUCAAAUGGAGUAGAA (((((((..((..((.(((((.......)))...((((((((.((((.....)))).(((..(((((.........)))))..)))....))))))))..))))..))..))))..))). ( -35.60) >consensus UCUUCCAAGUGGCAAUGCGGCACAGUGGGCCAGAUCGGCGCUGUAAUUAACUAUUAACAUGAGUG_CUCCGAUUGUGCCACAAAUGAGCAAUUGCCGAGAGCUUAUCAAAUGGAGUAGAA (((((((..(((((((((((((((((.((((.....)))..........(((.........))).....).)))))))).(....).)).))))))).((.....))...))))..))). (-32.62 = -32.46 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:09 2006