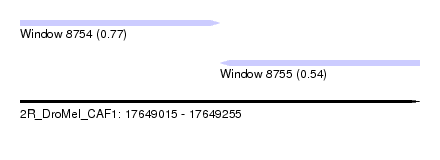
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,649,015 – 17,649,255 |
| Length | 240 |
| Max. P | 0.774347 |

| Location | 17,649,015 – 17,649,135 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.11 |
| Mean single sequence MFE | -25.96 |
| Consensus MFE | -16.34 |
| Energy contribution | -18.10 |
| Covariance contribution | 1.76 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.774347 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17649015 120 + 20766785 GUUUACCCCAUAGACGCUAUAGUUUUCUUAACUAAACUAGACUUGAGGCACUCAAGUUUUAAUCAUGCCCAGCCCAGAUCCCUGGCAGAAUCUAAUUACAUAUUUGGAUUCUGCCACUUU (((((.....)))))(((.((((((........))))))((((((((...))))))))............))).........(((((((((((((........))))))))))))).... ( -34.00) >DroSec_CAF1 2652 120 + 1 GUUUACCCCAUAGAAGCUAUAGCUUUCUUAACUUAACUAGACUUGAGGCACUCAAGUUUUAAUCAUGCCCAGCCCAGAUCCCUGGCAGAGUUUAAUUAUAUAUUUGGAUUCUGGCACUUU (((.........(((((....)))))........))).(((((((((...)))))))))......((((..(((.((....)))))(((((((((........))))))))))))).... ( -28.03) >DroSim_CAF1 8573 109 + 1 GUUUACCCCAUAGAAGCUAUAGCUUUCUUAACUUAACUAGACUUGAGGCACUCAAGUUUUAAUCAUGCCCAGCCCA-----------GAGUUUAAUUAUAUAUUUGGAUUCUGCCACUUU (((.........(((((....)))))........))).(((((((((...)))))))))...............((-----------((((((((........))))))))))....... ( -20.13) >DroEre_CAF1 2730 112 + 1 U--------UUUGAAGCUAUAUCUUUAUUAGUUAAGCCGGACUUGAGGCACUCAAGUUUUAAUCAAGUCUAACCCAGAUCCAUGGCGGAGUCUAAUUACAUAUGUGUGUUCUGCCACUUU .--------((((.(((((.........))))).....((((((((...((....)).....))))))))....))))....((((((((.(....(((....))).))))))))).... ( -23.30) >DroYak_CAF1 13962 112 + 1 U--------UUUGAGGCUCUGACUUUUUAAGCUUAACCAGACUGGAGGCACUCAAGUUUUAAUCAUGCCCAAUCCAGAUCCCUGGCAGAGUCUAAUUACAUAUUUGUGUUGUGCCACUUU .--------.....(((.(.(((...(((..(((..((((.(((((((((...............))))...)))))....))))..)))..)))..(((....))))))).)))..... ( -24.36) >consensus GUUUACCCCAUAGAAGCUAUAGCUUUCUUAACUUAACUAGACUUGAGGCACUCAAGUUUUAAUCAUGCCCAGCCCAGAUCCCUGGCAGAGUCUAAUUACAUAUUUGGAUUCUGCCACUUU ............(((((....)))))............(((((((((...))))))))).......................(((((((((((((........))))))))))))).... (-16.34 = -18.10 + 1.76)

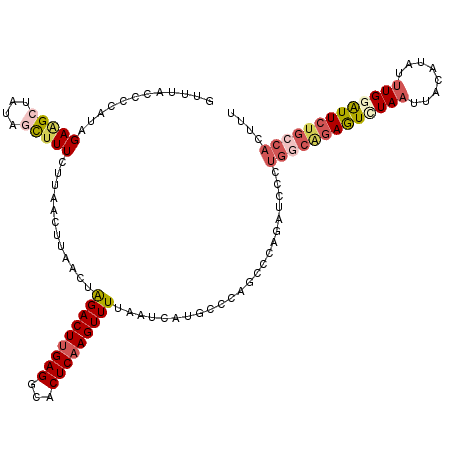
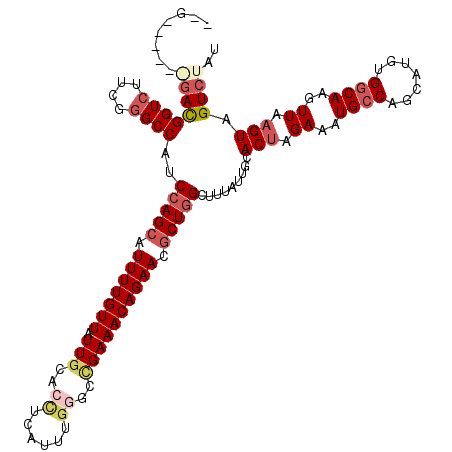
| Location | 17,649,135 – 17,649,255 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.01 |
| Mean single sequence MFE | -35.80 |
| Consensus MFE | -23.70 |
| Energy contribution | -25.62 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.537798 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17649135 120 - 20766785 AUGUACAUGGACGGUCUUCGGGCCAUCCAGCAUUUUGUUAUUGCACCUCAUUUUGGCUGAAACAGAACGCUGGCUUAAUUGCACUAGAAAUGCCAGCAUGUGGCAAGUUAAGUAGUCUAU ......((((((((((....))))..(((((.(((((((....((((.......)).)).))))))).))))).........(((.((..(((((.....)))))..)).))).)))))) ( -37.70) >DroSec_CAF1 2772 114 - 1 --G----GAGACGGUCUUCGGGCCAUCCAGCAUUUUGUUAUUGCACCUCACUUGAGCCGAAACAGAACGCUGGCUUUAUUGCACUAGAAAUGCCAGCAUGUGGCAUGUUAAGUAGUCUAU --.----.((((((((....))))..(((((.(((((((.(((...(((....))).)))))))))).))))).........(((.((.((((((.....)))))).)).))).)))).. ( -38.10) >DroSim_CAF1 8682 114 - 1 --G----GAGACGGUCUUCUUGCCAUCCAGCAUUUUGUUAUUGCACCUCAUUUGGGCCGAAACAGAACGCUGGCUUUAUUGCACCAGAAAUGCCAGCAUGUGGCAAGUUAAGUAGUCUAU --.----.((((...((((((((((((((((.(((((((...((.((......))))...))))))).)))))......(((..((....))...))).))))))))..)))..)))).. ( -38.50) >DroEre_CAF1 2842 108 - 1 --G------GACGGUCCUCGGGCCAUCGAGCAUUUUGUUAUUGCACUUCAUUUG----GAAACAGAACCCUGGCUAUAUUGCACUGGAAAUGCCUGCAUGCGGCAAGUUAAGUAGUCUAU --(------(((((((....)))).....(((...(((....((......((((----....))))......)).))).)))(((.((..((((.......))))..)).))).)))).. ( -27.00) >DroYak_CAF1 14074 112 - 1 --C------UAUGGUCUUCGGGCCAUCCAGCAUUUUGUUAUUGCACCUCAUUUGGUCCGAAACAGAACGCUGGCUAUAUUGCACUGGAAGUGGCUGCAUGUGGCAAGUUAAGUAGUCUAU --.------.((((((....)))))).((((.(((((((.(((.(((......))).)))))))))).))))(((((((.(((((......)).))))))))))................ ( -37.70) >consensus __G______GACGGUCUUCGGGCCAUCCAGCAUUUUGUUAUUGCACCUCAUUUGGGCCGAAACAGAACGCUGGCUUUAUUGCACUAGAAAUGCCAGCAUGUGGCAAGUUAAGUAGUCUAU ........((((((((....))))..(((((.(((((((.(((..((......))..)))))))))).))))).........(((.((..((((.......))))..)).))).)))).. (-23.70 = -25.62 + 1.92)

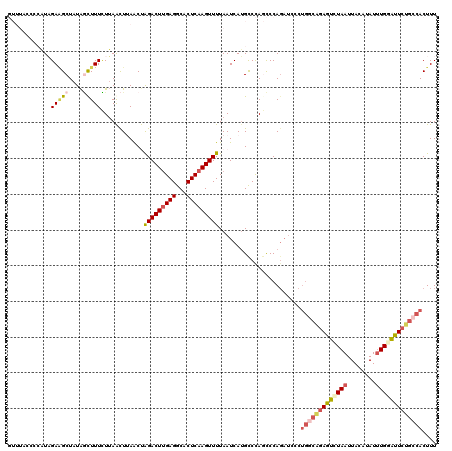
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:07 2006