| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,646,147 – 17,646,256 |
| Length | 109 |
| Max. P | 0.571670 |

| Location | 17,646,147 – 17,646,256 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.38 |
| Mean single sequence MFE | -24.41 |
| Consensus MFE | -16.57 |
| Energy contribution | -16.57 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.525466 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17646147 109 + 20766785 -AUAUCAUACUCAUAUGUAUGUGUGCAAAUGAUAUA----------UCUGUCAGUUGUCCAUCAUUGGCUAAGAUCAAAUCCGAAUCUAAGCCAUUUCGAGUUGAACAAACCCAAUCGCA -...(((.((((......(((((..(((.(((((..----------..))))).)))..)).)))(((((.(((((......).)))).)))))....)))))))............... ( -24.40) >DroSim_CAF1 5673 109 + 1 -AUAUCAUACUCAUAU----GUGUGCAAAUGAUAUAGAAGACUGGCUCUGUCAGCUGUCUAUCAAUGGCUAAGAUC------GAAUCUAAGCCAUUUCGAGUUGAACAAACCCAAUCGCA -(((((((...(((..----..)))...)))))))....((.(((...((((((((.......(((((((.((((.------..)))).)))))))...))))).)))...))).))... ( -25.20) >DroYak_CAF1 11003 110 + 1 UAUAUCAUUCACAUAU----GCAUGCAAAUGGUAUGGAAGACUGGUACUGUCAGCUGUCCAUCAUUGGCUAAGAUC------GAAACUAAGCCAUUUCGAGUUGAACAAACCCAAUCGCA (((((((((..(((..----..)))..)))))))))...((.(((...((((((((.........(((((.((...------....)).))))).....))))).)))...))).))... ( -23.64) >consensus _AUAUCAUACUCAUAU____GUGUGCAAAUGAUAUAGAAGACUGG_UCUGUCAGCUGUCCAUCAUUGGCUAAGAUC______GAAUCUAAGCCAUUUCGAGUUGAACAAACCCAAUCGCA .(((((((...(((((....)))))...))))))).............((((((((.........(((((.((((.........)))).))))).....))))).)))............ (-16.57 = -16.57 + 0.01)

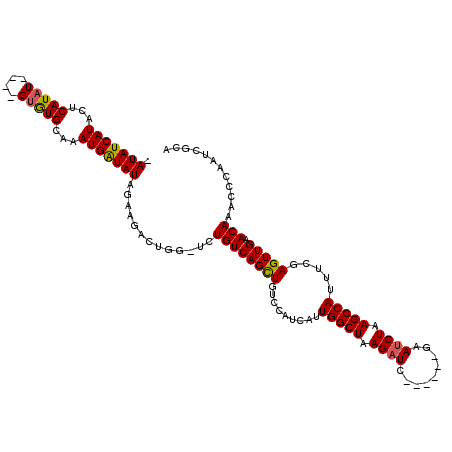
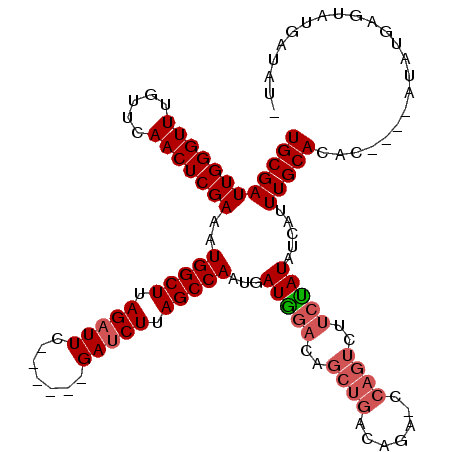
| Location | 17,646,147 – 17,646,256 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.38 |
| Mean single sequence MFE | -28.50 |
| Consensus MFE | -19.28 |
| Energy contribution | -21.17 |
| Covariance contribution | 1.89 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.571670 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17646147 109 - 20766785 UGCGAUUGGGUUUGUUCAACUCGAAAUGGCUUAGAUUCGGAUUUGAUCUUAGCCAAUGAUGGACAACUGACAGA----------UAUAUCAUUUGCACACAUACAUAUGAGUAUGAUAU- (((((.(((..(((((((..(((...(((((.(((((.......))))).))))).))))))))))(.....).----------....))).)))))..(((((......)))))....- ( -25.00) >DroSim_CAF1 5673 109 - 1 UGCGAUUGGGUUUGUUCAACUCGAAAUGGCUUAGAUUC------GAUCUUAGCCAUUGAUAGACAGCUGACAGAGCCAGUCUUCUAUAUCAUUUGCACAC----AUAUGAGUAUGAUAU- ...((((((.((((((....((..(((((((.((((..------.)))).)))))))....)).....)))))).))))))....(((((((...((...----...))...)))))))- ( -31.60) >DroYak_CAF1 11003 110 - 1 UGCGAUUGGGUUUGUUCAACUCGAAAUGGCUUAGUUUC------GAUCUUAGCCAAUGAUGGACAGCUGACAGUACCAGUCUUCCAUACCAUUUGCAUGC----AUAUGUGAAUGAUAUA .....(((((((.....)))))))..(((((.((....------...)).)))))...(((((..((((.......))))..)))))..((((..(((..----..)))..))))..... ( -28.90) >consensus UGCGAUUGGGUUUGUUCAACUCGAAAUGGCUUAGAUUC______GAUCUUAGCCAAUGAUGGACAGCUGACAGA_CCAGUCUUCUAUAUCAUUUGCACAC____AUAUGAGUAUGAUAU_ ((((((((((((.....)))))))..(((((.(((((.......))))).)))))...(((((..((((.......))))..))))).....)))))....................... (-19.28 = -21.17 + 1.89)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:04 2006