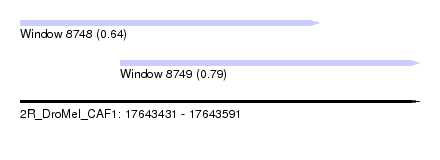
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,643,431 – 17,643,591 |
| Length | 160 |
| Max. P | 0.788824 |

| Location | 17,643,431 – 17,643,551 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.71 |
| Mean single sequence MFE | -32.63 |
| Consensus MFE | -31.02 |
| Energy contribution | -31.13 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639301 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
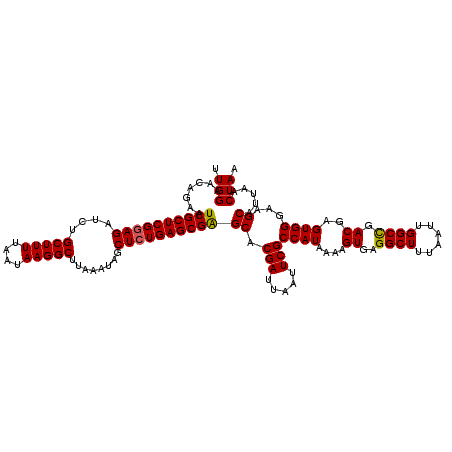
>2R_DroMel_CAF1 17643431 120 + 20766785 AAACUAUUAAGCGGAUUUGGCCAAUUCGGUAGCGGACAGAUUGGAACAGAAUCGCUCGGAGAUCUGCUUUUAAUAAGGCUUAAAUAGCUCUGAGCGAGCACGAUUAAUUCGCCAUAAAAG ..........(((((((((.(((((..(........)..)))))..)))).((((((((((....(((((....)))))........))))))))))..........)))))........ ( -33.50) >DroSim_CAF1 2935 119 + 1 A-ACUAUUAAGCGGAUUUGGCCAAUUCGGUAGCGAACAGAUUGGAACAGAAUCGCUCGGAGAUCUGCUUUUAAUAAGGCUUAAAUAACUCUGAGCGAGCACGAUUAAUUCGCCAUAAAAG .-........(((((((((.(((((..(........)..)))))..)))).((((((((((....(((((....)))))........))))))))))..........)))))........ ( -32.40) >DroYak_CAF1 8333 119 + 1 G-GCUAUUAAGCGGAUUUGGCCAAUUCGGUAGCGUACAGAUUGGAAAAGAAUCGCUCGUAGAUCUGCUUUUAAUAAGGCUUAAAUGGCUCUGAGCGGGCACGAUUAAUUCGCCAUAAAAG (-((.(((((.(((((((..(((((..(........)..)))))....)))))(((((((((.(((((((....)))))......)).)))..)))))).)).)))))..)))....... ( -32.00) >consensus A_ACUAUUAAGCGGAUUUGGCCAAUUCGGUAGCGAACAGAUUGGAACAGAAUCGCUCGGAGAUCUGCUUUUAAUAAGGCUUAAAUAGCUCUGAGCGAGCACGAUUAAUUCGCCAUAAAAG ..........(((((((...(((((..(........)..))))).......((((((((((....(((((....)))))........))))))))))........)))))))........ (-31.02 = -31.13 + 0.11)


| Location | 17,643,471 – 17,643,591 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -34.30 |
| Consensus MFE | -31.67 |
| Energy contribution | -31.57 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.788824 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17643471 120 + 20766785 UUGGAACAGAAUCGCUCGGAGAUCUGCUUUUAAUAAGGCUUAAAUAGCUCUGAGCGAGCACGAUUAAUUCGCCAUAAAAGUGAGGCUUUAAUUGGCUGACGAGUGGGAAGCAUUAACUAA .(((.......((((((((((....(((((....)))))........))))))))))((.(((.....)))((((....((.((.(.......).)).))..))))...)).....))). ( -33.60) >DroSim_CAF1 2974 120 + 1 UUGGAACAGAAUCGCUCGGAGAUCUGCUUUUAAUAAGGCUUAAAUAACUCUGAGCGAGCACGAUUAAUUCGCCAUAAAAGUGAGGCUUUAAUUGGCCGACGAGUGGGAAGCAUUAACUAA .(((.......((((((((((....(((((....)))))........))))))))))((.(((.....)))((((....((..((((......)))).))..))))...)).....))). ( -34.40) >DroYak_CAF1 8372 120 + 1 UUGGAAAAGAAUCGCUCGUAGAUCUGCUUUUAAUAAGGCUUAAAUGGCUCUGAGCGGGCACGAUUAAUUCGCCAUAAAAGUGUGGCUUUAAUUGGCCGACGAGUGGGAAGCAUUAACUAA ...........((((((((.....(((((.......((((.....))))..)))))(((.(((((((...((((((....)))))).)))))))))).)))))))).............. ( -34.90) >consensus UUGGAACAGAAUCGCUCGGAGAUCUGCUUUUAAUAAGGCUUAAAUAGCUCUGAGCGAGCACGAUUAAUUCGCCAUAAAAGUGAGGCUUUAAUUGGCCGACGAGUGGGAAGCAUUAACUAA .(((.......((((((((((....(((((....)))))........))))))))))((.(((.....)))((((....((..((((......)))).))..))))...)).....))). (-31.67 = -31.57 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:01 2006