
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,641,516 – 17,641,716 |
| Length | 200 |
| Max. P | 0.888167 |

| Location | 17,641,516 – 17,641,636 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.50 |
| Mean single sequence MFE | -25.90 |
| Consensus MFE | -14.13 |
| Energy contribution | -14.69 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888167 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17641516 120 - 20766785 ACAUCUGUUAGAUGGCGCCACUAAGCCAAGCUUAUCUUUAAGCUAAGACAGCGAAAAAGCUAGUUUUAAAUUUAAGCUUACAUCAAACCCGUUUCACCUUAGCGCAAACAUUAAGAGCAU .((((.....))))((((...(((((...(((((....)))))(((((((((......))).)))))).......)))))....(((....))).......))))............... ( -24.50) >DroSim_CAF1 938 114 - 1 CCAUCUGUUAGAUGGCGCCACUAAGCCAAGCUUGUCUUUAAGCUGAGACAGCAAAAAAGCUAGUUUCAAAUGAAAGCUUACAUCAAGCC-----CACCUUAACGCUAACAUUAAAA-AAU .....((((((.((((........)))).(((((....((((((((((((((......))).))))).......))))))...))))).-----..........))))))......-... ( -29.41) >DroYak_CAF1 6267 114 - 1 CCAUCUGUUAGAUGGCGCCACUUACCCGAGCUUGUCUUUUAGCUGAGACAACUAAAAAGCUAGUUUAAAAUUAAAGCUUAAUUUAAACCCGUUUGAAU------CGAGCACUACAAAAAA (((((.....)))))............(((((.(..((((((.((...)).))))))..).))))).........(((..(((((((....)))))))------..)))........... ( -23.80) >consensus CCAUCUGUUAGAUGGCGCCACUAAGCCAAGCUUGUCUUUAAGCUGAGACAGCAAAAAAGCUAGUUUAAAAUUAAAGCUUACAUCAAACCCGUUUCACCUUA_CGCAAACAUUAAAA_AAU ....(((((...((((........))))((((((....))))))..))))).....(((((.............)))))......................................... (-14.13 = -14.69 + 0.56)

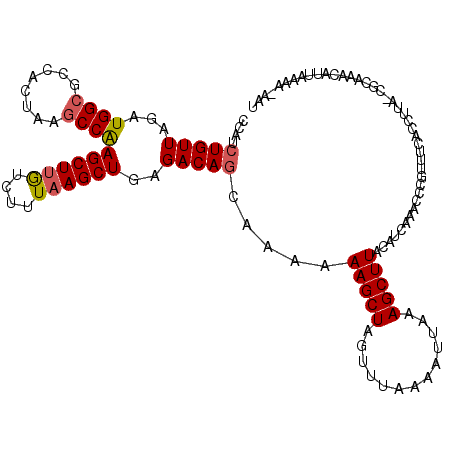
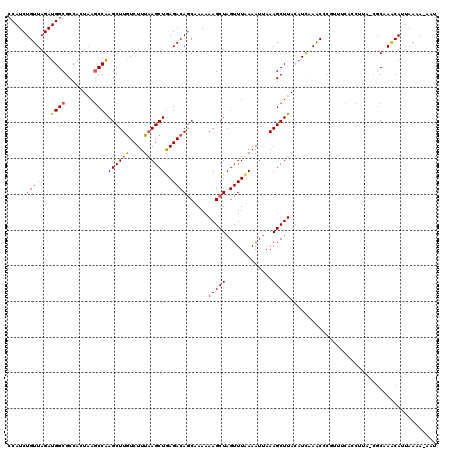
| Location | 17,641,556 – 17,641,676 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.67 |
| Mean single sequence MFE | -32.13 |
| Consensus MFE | -24.09 |
| Energy contribution | -24.87 |
| Covariance contribution | 0.79 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.75 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17641556 120 + 20766785 UAAGCUUAAAUUUAAAACUAGCUUUUUCGCUGUCUUAGCUUAAAGAUAAGCUUGGCUUAGUGGCGCCAUCUAACAGAUGUUCUGCUCAAUUUUCAUGACCAACAGAUGGCGUAGCUUAGG ((((((.....................(((((.((.((((((....)))))).))..)))))(((((((((..(((.....))).(((.......))).....))))))))))))))).. ( -32.90) >DroSim_CAF1 972 120 + 1 UAAGCUUUCAUUUGAAACUAGCUUUUUUGCUGUCUCAGCUUAAAGACAAGCUUGGCUUAGUGGCGCCAUCUAACAGAUGGUUUGCUCAAUUUUCAUGCCCAACAGAUGGCGUAACUAUGG ((((((......(((.((.(((......))))).)))((((......))))..))))))((.(((((((((....(.(((...((...........))))).)))))))))).))..... ( -30.90) >DroYak_CAF1 6301 120 + 1 UAAGCUUUAAUUUUAAACUAGCUUUUUAGUUGUCUCAGCUAAAAGACAAGCUCGGGUAAGUGGCGCCAUCUAACAGAUGGUCUGCUCAAAUUUCUUAGCCAACAGAUGGCGUAAGUGAGU ...(((.....((((..(.(((((((((((((...))))))))....))))).)..)))).)))((((((.....))))))..(((((.....((((((((.....)))).))))))))) ( -32.60) >consensus UAAGCUUUAAUUUAAAACUAGCUUUUUAGCUGUCUCAGCUUAAAGACAAGCUUGGCUUAGUGGCGCCAUCUAACAGAUGGUCUGCUCAAUUUUCAUGACCAACAGAUGGCGUAACUAAGG (((((((............(((......)))((((........)))))))))))....(((.(((((((((....(.(((((..............))))).)))))))))).))).... (-24.09 = -24.87 + 0.79)

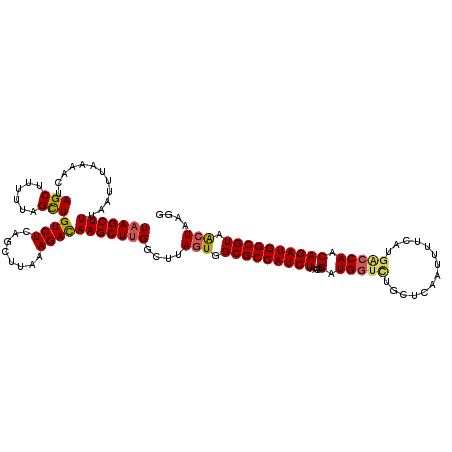
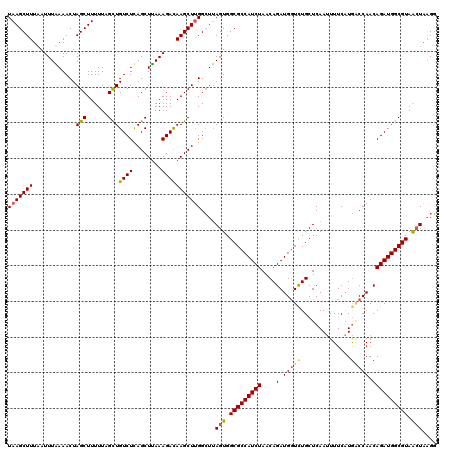
| Location | 17,641,556 – 17,641,676 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.67 |
| Mean single sequence MFE | -32.79 |
| Consensus MFE | -24.62 |
| Energy contribution | -24.73 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.872641 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17641556 120 - 20766785 CCUAAGCUACGCCAUCUGUUGGUCAUGAAAAUUGAGCAGAACAUCUGUUAGAUGGCGCCACUAAGCCAAGCUUAUCUUUAAGCUAAGACAGCGAAAAAGCUAGUUUUAAAUUUAAGCUUA ..((((((.(((((((((....(((.......)))((((.....)))))))))))))............(((((....)))))(((((((((......))).))))))......)))))) ( -34.50) >DroSim_CAF1 972 120 - 1 CCAUAGUUACGCCAUCUGUUGGGCAUGAAAAUUGAGCAAACCAUCUGUUAGAUGGCGCCACUAAGCCAAGCUUGUCUUUAAGCUGAGACAGCAAAAAAGCUAGUUUCAAAUGAAAGCUUA ...((((..(((((((((.(((..................))).....)))))))))..))))(((..((((((....))))))((((((((......))).)))))........))).. ( -34.17) >DroYak_CAF1 6301 120 - 1 ACUCACUUACGCCAUCUGUUGGCUAAGAAAUUUGAGCAGACCAUCUGUUAGAUGGCGCCACUUACCCGAGCUUGUCUUUUAGCUGAGACAACUAAAAAGCUAGUUUAAAAUUAAAGCUUA .....((((.((((.....)))))))).....((.((.(.(((((.....)))))))))).......(((((((((((......)))))(((((......)))))........)))))). ( -29.70) >consensus CCUAAGUUACGCCAUCUGUUGGCCAUGAAAAUUGAGCAGACCAUCUGUUAGAUGGCGCCACUAAGCCAAGCUUGUCUUUAAGCUGAGACAGCAAAAAAGCUAGUUUAAAAUUAAAGCUUA .........((((((((.......((....))..(((((.....)))))))))))))..........(((((((((((......)))))(((......)))............)))))). (-24.62 = -24.73 + 0.12)

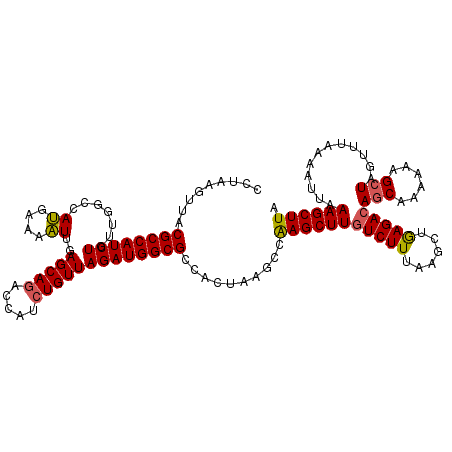

| Location | 17,641,596 – 17,641,716 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.86 |
| Mean single sequence MFE | -25.69 |
| Consensus MFE | -14.88 |
| Energy contribution | -14.77 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.658744 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17641596 120 - 20766785 AAAACAUAAAAUAUACAUGACAGCUAUCUACGUCCGCUGUCCUAAGCUACGCCAUCUGUUGGUCAUGAAAAUUGAGCAGAACAUCUGUUAGAUGGCGCCACUAAGCCAAGCUUAUCUUUA ..................((((((...........)))))).((((((..((((((((....(((.......)))((((.....))))))))))))((......))..))))))...... ( -30.20) >DroSim_CAF1 1012 100 - 1 -------AAACUAUACAUGACAGC-------------UGUCCAUAGUUACGCCAUCUGUUGGGCAUGAAAAUUGAGCAAACCAUCUGUUAGAUGGCGCCACUAAGCCAAGCUUGUCUUUA -------...........((((((-------------(...(.((((..(((((((((.(((..................))).....)))))))))..)))).)...))).)))).... ( -27.17) >DroYak_CAF1 6341 112 - 1 AAAACAUAAUCUAUUCAUGUCACGUAU--------AAUACACUCACUUACGCCAUCUGUUGGCUAAGAAAUUUGAGCAGACCAUCUGUUAGAUGGCGCCACUUACCCGAGCUUGUCUUUU ...........................--------......(((.((((.((((.....)))))))).....((.((.(.(((((.....)))))))))).......))).......... ( -19.70) >consensus AAAACAUAAACUAUACAUGACAGCUAU__________UGUCCUAAGUUACGCCAUCUGUUGGCCAUGAAAAUUGAGCAGACCAUCUGUUAGAUGGCGCCACUAAGCCAAGCUUGUCUUUA .................................................((((((((.......((....))..(((((.....)))))))))))))....................... (-14.88 = -14.77 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:58 2006