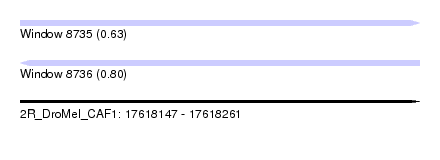
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 17,618,147 – 17,618,261 |
| Length | 114 |
| Max. P | 0.795919 |

| Location | 17,618,147 – 17,618,261 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 80.12 |
| Mean single sequence MFE | -50.58 |
| Consensus MFE | -34.64 |
| Energy contribution | -35.62 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.629156 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17618147 114 + 20766785 GUGCCUGGUCACCGCCGGACGCUGCUGUCCGUUGGCCAUCGCCAGUUGAUUCAAGCCCGGUGGCUUCGGAGCAAAAUUGGGCUUGCCAAAGCUGAUGGCCGGCUUCAUCG---GCAG (((.(((((....))))).)))(((((...((((((((((((...(((...((((((((((.(((....)))...)))))))))).))).)).)))))))))).....))---))). ( -52.90) >DroVir_CAF1 8289 114 + 1 AUGCCGCGCCACCGCCGGCCGCUGAUGCCCGUUGGCGAGCGCCAGUUGCUGCAGGCCCGGCGGCUUUGGCGCAAAGUUUGGCUUGCCAAACCCAAUGGCCGGCUUCUGUU---GCUG ..((((((((((((((((((((....((...(((((....)))))..)).)).)).)))))))...))))))...((((((....)))))).....)))((((.......---)))) ( -55.30) >DroPse_CAF1 12312 114 + 1 GUGCCGCGUCACCGCCGGACGCUGCUGCCCGUUAACCAGAAUCAACUGCUGGAGCCCCGGCGGCUUUGGUGCAAAGUUCGGCUUGCCAAAGCUGAUGGCCGGCUUCUGCU---GCUG ..(((((..(((((((((..(((.(.((..(((..........))).)).).))).)))))))...))..)).....(((((((....))))))).)))((((....)))---)... ( -43.30) >DroMoj_CAF1 23819 114 + 1 AUGUCGUGCCACCGCUGGACGCUGAUGCCCAUUGGCUAACGCCAGCUGCUGCAGCCCCGGCGGCUUUGGCGCAAAAUUUGGCUUGCCAAAGCCAAUUGCUGGCUUCUGUU---GCUG .....(((((((((((((..((((..((...(((((....)))))..))..)))).)))))))...)))))).......(((..((..((((((.....))))))..)).---))). ( -50.00) >DroAna_CAF1 6961 117 + 1 GUGCCUGGUCACCGCCGGACGUUGCUGGCCGUUGGCCAAGGCCAGUUGCUGCAGCCCCGGCGGCUUCGGCGCGAAGUUCGGUUUCCCGAAGCUAAUCGCCGGCUUCGUGGUGUGCUG (((((.((...(((((((..(((((.(((..(((((....)))))..)))))))).)))))))..)))))))......((((..(((((((((.......))))))).))...)))) ( -58.70) >DroPer_CAF1 12301 114 + 1 GUGCCGCGUCACCGCCGGACGCUGCUGCCCGUUAACCAGAAUCAACUGCUGGAGCCCCGGCGGCUUUGGUGCAAAGUUCGGCUUGCCAAAGCUGAUGGCCGGCUUCUGCU---GCUG ..(((((..(((((((((..(((.(.((..(((..........))).)).).))).)))))))...))..)).....(((((((....))))))).)))((((....)))---)... ( -43.30) >consensus GUGCCGCGUCACCGCCGGACGCUGCUGCCCGUUGGCCAGCGCCAGCUGCUGCAGCCCCGGCGGCUUUGGCGCAAAGUUCGGCUUGCCAAAGCUAAUGGCCGGCUUCUGCU___GCUG ..((((.((((..(((((..((((....(.((((((....)))))).)...)))).)))))(((((((((((........))..)))))))))..)))))))).............. (-34.64 = -35.62 + 0.98)

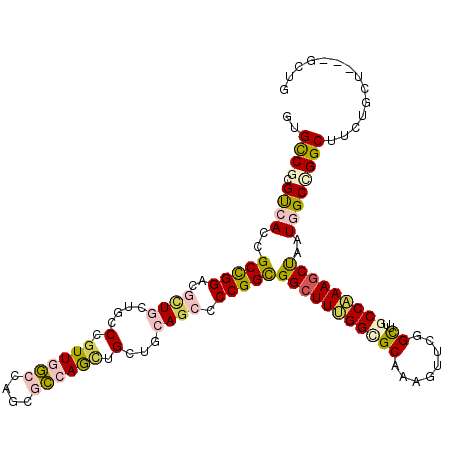
| Location | 17,618,147 – 17,618,261 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 80.12 |
| Mean single sequence MFE | -49.73 |
| Consensus MFE | -32.47 |
| Energy contribution | -32.92 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795919 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 17618147 114 - 20766785 CUGC---CGAUGAAGCCGGCCAUCAGCUUUGGCAAGCCCAAUUUUGCUCCGAAGCCACCGGGCUUGAAUCAACUGGCGAUGGCCAACGGACAGCAGCGUCCGGCGGUGACCAGGCAC .(((---(......((((((((((.((((((.(((((((......(((....)))....)))))))...)))..))))))))))...((((......)))))))(....)..)))). ( -50.10) >DroVir_CAF1 8289 114 - 1 CAGC---AACAGAAGCCGGCCAUUGGGUUUGGCAAGCCAAACUUUGCGCCAAAGCCGCCGGGCCUGCAGCAACUGGCGCUCGCCAACGGGCAUCAGCGGCCGGCGGUGGCGCGGCAU ....---.......(((.......((((((((....)))))))).(((((...((((((((.((((..((...((((....))))....))..))).).)))))))))))))))).. ( -56.00) >DroPse_CAF1 12312 114 - 1 CAGC---AGCAGAAGCCGGCCAUCAGCUUUGGCAAGCCGAACUUUGCACCAAAGCCGCCGGGGCUCCAGCAGUUGAUUCUGGUUAACGGGCAGCAGCGUCCGGCGGUGACGCGGCAC ....---.......(((((.((((.(((((((...((........)).))))))).(((((((((.(.((.((((((....))))))..)).).))).)))))))))).).)))).. ( -44.40) >DroMoj_CAF1 23819 114 - 1 CAGC---AACAGAAGCCAGCAAUUGGCUUUGGCAAGCCAAAUUUUGCGCCAAAGCCGCCGGGGCUGCAGCAGCUGGCGUUAGCCAAUGGGCAUCAGCGUCCAGCGGUGGCACGACAU ....---.......((((((....(((((((((..((........)))))))))))((....)).......))))))(((.((((.(((((......)))))....))))..))).. ( -46.90) >DroAna_CAF1 6961 117 - 1 CAGCACACCACGAAGCCGGCGAUUAGCUUCGGGAAACCGAACUUCGCGCCGAAGCCGCCGGGGCUGCAGCAACUGGCCUUGGCCAACGGCCAGCAACGUCCGGCGGUGACCAGGCAC ..............(((((((...((.(((((....)))))))...))))...(((((((((((((..((...((((....))))...))))))....))))))))).....))).. ( -56.60) >DroPer_CAF1 12301 114 - 1 CAGC---AGCAGAAGCCGGCCAUCAGCUUUGGCAAGCCGAACUUUGCACCAAAGCCGCCGGGGCUCCAGCAGUUGAUUCUGGUUAACGGGCAGCAGCGUCCGGCGGUGACGCGGCAC ....---.......(((((.((((.(((((((...((........)).))))))).(((((((((.(.((.((((((....))))))..)).).))).)))))))))).).)))).. ( -44.40) >consensus CAGC___AGCAGAAGCCGGCCAUCAGCUUUGGCAAGCCGAACUUUGCGCCAAAGCCGCCGGGGCUGCAGCAACUGGCGCUGGCCAACGGGCAGCAGCGUCCGGCGGUGACGCGGCAC ..............(((.((....((.(((((....)))))))..))......(((((((((((((..((...((((....))))....))..)))).))))))))).....))).. (-32.47 = -32.92 + 0.45)
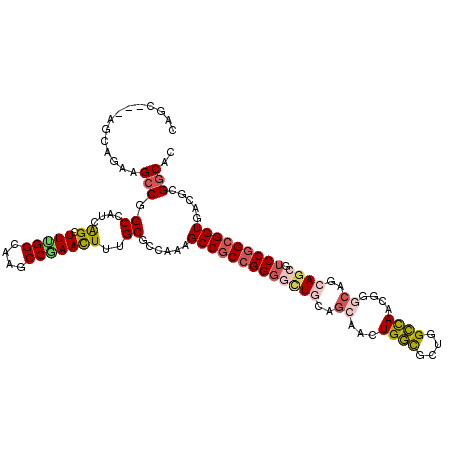
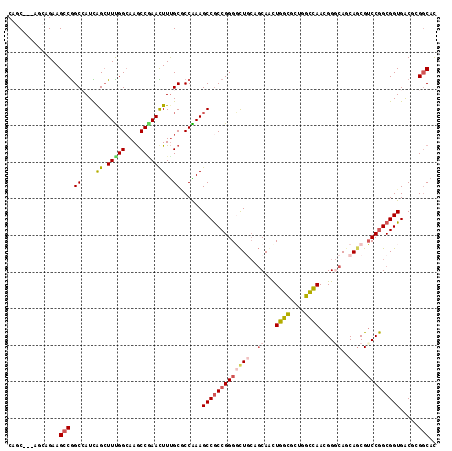
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:48 2006